Coxiella burnetii type IVB secretion system region I genes are expressed early during the infection of host cells - PubMed (original) (raw)
Coxiella burnetii type IVB secretion system region I genes are expressed early during the infection of host cells
John K Morgan et al. FEMS Microbiol Lett. 2010 Oct.
Abstract
Analysis of the Coxiella burnetii RSA 493 (Nine Mile phase I strain) genome revealed ORFs with significant homology to the type IVB secretion system (T4BSS) of Legionella pneumophila. The T4BSS genes exist primarily at two loci, designated regions I (RI) and II. In C. burnetii, little is known about the T4BSS regions and the role they play in establishing and/or maintaining infection. Coxiella burnetii T4BSS RI contains genes arranged in three linkage groups: (1) icmW→CBU1651→icmX, (2) icmV→dotA→CBU1647, and (3) icmT→icmS→dotD→dotC→dotB→CBU1646. We used reverse transcriptase (RT)-PCR to demonstrate transcriptional linkage within the groups, and that icmX, icmV, and icmT are transcribed de novo by 8 h post infection (hpi). We then examined the transcript levels for icmX, icmW, icmV, dotA, dotB, and icmT during the first 24 h of an infection using quantitative RT-PCR. The expression initially increased for each gene, followed by a decrease at 24 hpi. Subsequently, we analyzed IcmT protein levels during infection and determined that the expression increases significantly from 8 to 24 hpi and then remains relatively constant. These data demonstrate temporal changes in the RNA of several C. burnetii T4SS RI homologs and the IcmT protein. These changes correspond to early stages of the C. burnetii infectious cycle.
© 2010 Federation of European Microbiological Societies. Published by Blackwell Publishing Ltd. All rights reserved.
Figures
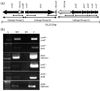
Fig. 1. Coxiella burnetii T4BSS Region I gene linkage
(a) Physical map of C. burnetii T4BSS RI. Solid arrows represent T4BSS homologs. Open arrows represent non-T4BSS ORFs. Gene designations are indicated above each ORF. Diamond-ended lines indicate the position of primers and DNA products that would result from RT-PCR amplification. (b) Agarose gel image(s) showing RT-PCR products corresponding to co-transcribed genes from Linkage Group i, ii, and iii (correlating to the diamond tipped lines in (a). L, 100 bp DNA ladder (size designated on left). +RT, with reverse transcriptase. −RT, without reverse transcriptase. C, DNA control.
Fig. 2. RT-PCR detection of C. burnetii T4BSS transcripts, icmT, icmV, and icmW
RNA template was isolated at 8 hpi from rifampicin-treated (+Rif) and mock-treated (−Rif) cells. L, 100 bp DNA ladder (size designated on left). +RT/−Rif, with reverse transcriptase and mock-treated. −RT/−Rif, without reverse transcriptase and mock-treated. +RT/+Rif, with reverse transcriptase and rifampin-treated. −RT/+Rif, without reverse transcriptase and rifampin-treated. C, DNA control.
Fig. 3. C. burnetii icmX, icmW, icmV, dotA, dotB, and icmT transcript levels during the early stages of infection
(a–f) Fold changes in mRNA levels relative to 0 hpi. An equal amount of total RNA from each sample was analyzed by RT-qPCR. The time (in hpi) when total RNA was harvested is indicated below the X-axis. Results represent the mean of three biological samples with no fewer than three technical replicates of each sample. Standard error bars represent the combined standard error of the mean (S.E.M.) per time point.
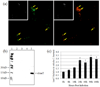
Fig. 4. Relative IcmT expression over a time course of infection
(a) Representative dual channel IFA micrograph (400× magnification) images of C. burnetii infected Vero cells 24 hpi. Left panel - Alexa® 488 indicates RαIcmT antibody binding. Right panel - Alexa® 555 indicates Guinea pig anti-whole C. burnetii antibody. Inset images are 8 bit (gray scale) conversions of the color images. Representative Regions of Interest (ROI) are shown (arrows). (b) Immunoblot using RαIcmT to probe total protein from purified C. burnetii (lane 2), Vero cells (lane 3), blank lane (lane 4), and purified recombinant IcmT (rIcmT, lane 5). Lane 1 contains the protein size ladder. (c) Pixel density ratios of IcmT to whole C. burnetii (488:555) from digital images captured at 0, 8, 16, 24, 48, 96, and 168 hpi were converted to quotient values relative to 0 hpi. Error bars represent the S.E.M. for each respective quotient value.
Similar articles
- Dependency of Coxiella burnetii Type 4B Secretion on the Chaperone IcmS.
Larson CL, Beare PA, Heinzen RA. Larson CL, et al. J Bacteriol. 2019 Nov 5;201(23):e00431-19. doi: 10.1128/JB.00431-19. Print 2019 Dec 1. J Bacteriol. 2019. PMID: 31501284 Free PMC article. - Polar localization of the Coxiella burnetii type IVB secretion system.
Morgan JK, Luedtke BE, Shaw EI. Morgan JK, et al. FEMS Microbiol Lett. 2010 Apr;305(2):177-83. doi: 10.1111/j.1574-6968.2010.01926.x. Epub 2010 Feb 9. FEMS Microbiol Lett. 2010. PMID: 20199576 Free PMC article. - Identification of Type 4B Secretion System Substrates That Are Conserved among Coxiella burnetii Genomes and Promote Intracellular Growth.
Larson CL, Pullman W, Beare PA, Heinzen RA. Larson CL, et al. Microbiol Spectr. 2023 Jun 15;11(3):e0069623. doi: 10.1128/spectrum.00696-23. Epub 2023 May 18. Microbiol Spectr. 2023. PMID: 37199620 Free PMC article. - Coxiella burnetii as a useful tool to investigate bacteria-friendly host cell compartments.
Pechstein J, Schulze-Luehrmann J, Lührmann A. Pechstein J, et al. Int J Med Microbiol. 2018 Jan;308(1):77-83. doi: 10.1016/j.ijmm.2017.09.010. Epub 2017 Sep 14. Int J Med Microbiol. 2018. PMID: 28935173 Review. - Coxiella burnetii secretion systems.
McDonough JA, Newton HJ, Roy CR. McDonough JA, et al. Adv Exp Med Biol. 2012;984:171-97. doi: 10.1007/978-94-007-4315-1_9. Adv Exp Med Biol. 2012. PMID: 22711632 Review.
Cited by
- Effector protein translocation by the Coxiella burnetii Dot/Icm type IV secretion system requires endocytic maturation of the pathogen-occupied vacuole.
Newton HJ, McDonough JA, Roy CR. Newton HJ, et al. PLoS One. 2013;8(1):e54566. doi: 10.1371/journal.pone.0054566. Epub 2013 Jan 17. PLoS One. 2013. PMID: 23349930 Free PMC article. - Immunisation with purified Coxiella burnetii phase I lipopolysaccharide confers partial protection in mice independently of co-administered adenovirus vectored vaccines.
Dold C, Zhu H, Silva-Reyes L, Blackwell L, Linder A, Bewley K, Godwin K, Fotheringham S, Charlton S, Kim YC, Pollard AJ, Rollier CS. Dold C, et al. Vaccine. 2023 May 5;41(19):3047-3057. doi: 10.1016/j.vaccine.2023.04.012. Epub 2023 Apr 8. Vaccine. 2023. PMID: 37037709 Free PMC article. - The Coxiella Burnetii type IVB secretion system (T4BSS) component DotA is released/secreted during infection of host cells and during in vitro growth in a T4BSS-dependent manner.
Luedtke BE, Mahapatra S, Lutter EI, Shaw EI. Luedtke BE, et al. Pathog Dis. 2017 Jun 1;75(4):ftx047. doi: 10.1093/femspd/ftx047. Pathog Dis. 2017. PMID: 28449081 Free PMC article. - Major differential gene regulation in Coxiella burnetii between in vivo and in vitro cultivation models.
Kuley R, Bossers-deVries R, Smith HE, Smits MA, Roest HI, Bossers A. Kuley R, et al. BMC Genomics. 2015 Nov 16;16:953. doi: 10.1186/s12864-015-2143-7. BMC Genomics. 2015. PMID: 26572556 Free PMC article. - Coxiella burnetii replicates in Galleria mellonella hemocytes and transcriptome mapping reveals in vivo regulated genes.
Kovacs-Simon A, Metters G, Norville I, Hemsley C, Titball RW. Kovacs-Simon A, et al. Virulence. 2020 Dec;11(1):1268-1278. doi: 10.1080/21505594.2020.1819111. Virulence. 2020. PMID: 32970966 Free PMC article.
References
- Berger KH, Isberg RR. Two distinct defects in intracellular growth complemented by a single genetic locus in Legionella pneumophila. Mol Microbiol. 1993;7:7–19. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources