Moenomycin family antibiotics: chemical synthesis, biosynthesis, and biological activity - PubMed (original) (raw)
Review
. 2010 Nov;27(11):1594-617.
doi: 10.1039/c001461n. Epub 2010 Aug 23.
Affiliations
- PMID: 20730219
- PMCID: PMC2987538
- DOI: 10.1039/c001461n
Review
Moenomycin family antibiotics: chemical synthesis, biosynthesis, and biological activity
Bohdan Ostash et al. Nat Prod Rep. 2010 Nov.
Abstract
The review (with 214 references cited) is devoted to moenomycins, the only known group of antibiotics that directly inhibit bacterial peptidoglycan glycosytransferases. Naturally occurring moenomycins and chemical and biological approaches to their derivatives are described. The biological properties of moenomycins and plausible mechanisms of bacterial resistance to them are also covered here, portraying a complete picture of the chemistry and biology of these fascinating natural products
Figures
Fig. 1
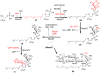
Moenomycin and its target. (A) Structures of bacterial cell wall-active antibiotics moenomycin A (1), penicllin G, and vancomycin. The building blocks of 1 are marked with capital letters A-H (red). (B) The extracellular steps of bacterial peptidoglycan biosynthesis and sites of antibiotic inhibition. Penicillin binding proteins (PBPs) are shown with both peptidoglycan glycosyltransferase (PGT) and transpeptidase (TP) domains. Black rectangles indicate the lipid carrier, undecaprenyl pyrophosphate. The sites of antibiotic action are indicated by red lines; the green arrow indicates the PGT reaction.
Fig. 2
Structures of naturally occuring moenomycins, as judged from MS and NMR experiments (except for 6 and 7, which are deduced only from MS analysis). The molecular mass of each compound (anion) is indicated (rightmost column).
Fig. 3
Di- and trisaccharide fragments and analogs of 1 obtained through degradation and chemical synthesis.
Fig. 4
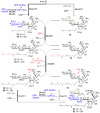
Synthetic moenomycins that resulted from modification or replacement of A ring.
Fig. 5
Genetic organization of moe clusters 1 and 2. Red bidirectional arrows indicate moe genes that are likely to arise from duplication event.
Fig. 6
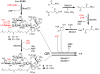
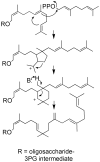
Early steps of moenomycin biosynthesis. Solid arrows indicate the order of moenomycin assembly. Dashed line indicates product 84 accumulated as a result of loss of moeK5 gene function. Structural components shown in red indicate the positions of new moieties in the intermediates.
Fig. 7
Sequence of reactions leading from disaccharide precursor 71 to tetrasaccharide 81/82. Structural components shown in red, green and blue indicate the positions of new moieties in the intermediates.
Fig. 8
Final steps of moenomycin biosynthesis. The sequence of reactions leading from pentasaccharide precursors to 1 is proposed, but may involve other intermediates and/or enzymes.
Fig. 9
Gene clusters that potentially may direct the production of phosphoglycolipid secondary metabolites.
Fig. 10
Summary of SAR studies around MmA scaffold. Functional groups of oligosaccharide-3PG moiety of 1 crucial for antibiotic activity are shown in dark orange. Grey shade indicates minimal in vivo pharmacophore 1 of MmA (30); another recently identified minimal pharmacophore 2 (72) contains D ring (shaded in yellow) instead of C ring. Key modifications of lipid and chromophore parts of MmA are listed on both sides of 1 and their relative antistaphylococcal activity is mentioned.
Fig. 11
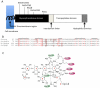
PGT structure and inhibition. A. Topology diagram depicting the arrangement of domains for a typical class A bifunctional pencillin-binding protein. The conserved residues of the PGT domain (signature motifs) are shown above the diagram. This domain arrangement is conserved in the monofunctional PGTs, but the linker region and TP domain are absent B. Multiple sequence alignment of a portion of PGT domains of S. aureus Mtg (Sa-mtg), PBP2 (Sa-pbp2), A. aeolicus PBP1a (Aa-pbp1a) and E. coli PBP1b (Ec-pbp1b). Five signature motifs are shown in red and lined above the alignment. For the sake of clarity, two aminoacid stretches are not shown in the alignment (marked with square brackets). Residues forming potential interactions with moenomycin pharmacophore are highlighted. C. A schematic view of interaction between pharmacophore (E-F-G-H units) and PGT. Signature motif aminoacids that contact 1 in at least 3 out of 4 structures are marked with filled magenta ovals; numbering is taken from 1:Mtg cocomplex. Other conservative residues are indicated by green ovals.
Fig. 12
Structural details of interaction between moenomycin and PGT and implications for PGT mechanism. A. Close-up view of surface representation of A. aeolicus Pbp1a co-complexed with neryl-moenomycin (55). The highly conserved residues of the 5 PGT motifs (colored magenta) are located in the active site cleft where 55 binds. Enzyme consists of a smaller lobe (closer to membrane) and a larger lobe. The lipid tail portion points down towards the proposed membrane interface. The 3-PG and pentasaccharide moieties extend upwards along the long groove. B. Working model for PGT-catalyzed reaction. Growing chain occupies donor substrate site and reacts with incoming lipid II unit (acceptor). The outer helix and the loop in the smaller lobe of PGT are mobile and help transfer the reaction product to donor site for next round of polymerization. Through binding to the donor site, moenomycins may prevent the initiation of polymerization or elongation of the first product, lipid IV (tetrasaccharide).
Fig. 13
Scheme 1
Synthesis of 16
Scheme 2
Moenomycins with modified lipid moiety.
Scheme 3
The total synthesis of 1.
Scheme 4
Proposed mechanism for rearrangement during biosynthesis of lipid portion of 1.
Similar articles
- Differential antibacterial activity of moenomycin analogues on gram-positive bacteria.
Goldman RC, Baizman ER, Branstrom AA, Longley CB. Goldman RC, et al. Bioorg Med Chem Lett. 2000 Oct 16;10(20):2251-4. doi: 10.1016/s0960-894x(00)00443-1. Bioorg Med Chem Lett. 2000. PMID: 11055331 - GERI-155, a new macrolide antibiotic related to chalcomycin.
Kim SD, Ryoo IJ, Kim CJ, Kim WG, Kim JP, Kong JY, Koshino H, Uramoto M, Yoo ID. Kim SD, et al. J Antibiot (Tokyo). 1996 Sep;49(9):955-7. doi: 10.7164/antibiotics.49.955. J Antibiot (Tokyo). 1996. PMID: 8931736 No abstract available. - Novel antibiotics SF2738A, B and C, and their analogs produced by Streptomyces sp.
Gomi S, Amano S, Sato E, Miyadoh S, Kodama Y. Gomi S, et al. J Antibiot (Tokyo). 1994 Dec;47(12):1385-94. doi: 10.7164/antibiotics.47.1385. J Antibiot (Tokyo). 1994. PMID: 7844033 - Glycosyltransferases involved in the biosynthesis of biologically active natural products that contain oligosaccharides.
Luzhetskyy A, Vente A, Bechthold A. Luzhetskyy A, et al. Mol Biosyst. 2005 Jul;1(2):117-26. doi: 10.1039/b503215f. Epub 2005 Jun 23. Mol Biosyst. 2005. PMID: 16880973 Review. - The molecular biology of moenomycins: towards novel antibiotics based on inhibition of bacterial peptidoglycan glycosyltransferases.
Ostash B, Doud E, Fedorenko V. Ostash B, et al. Biol Chem. 2010 May;391(5):499-504. doi: 10.1515/BC.2010.053. Biol Chem. 2010. PMID: 20302515 Review.
Cited by
- Discovery of new AMR drugs targeting modulators of antimicrobial activity using in vivo silkworm screening systems.
Tabuchi F, Mikami K, Miyauchi M, Sekimizu K, Miyashita A. Tabuchi F, et al. J Antibiot (Tokyo). 2024 Nov 14. doi: 10.1038/s41429-024-00788-2. Online ahead of print. J Antibiot (Tokyo). 2024. PMID: 39543333 Review. - Flavomycin restores colistin susceptibility in multidrug-resistant Gram-negative bacteria.
Huang Y, Zhu Y, Yue H-Y, Liu Y-Y, Deng L-M, Lv L, Wang C, Yang J, Liu J-H. Huang Y, et al. mSystems. 2024 Jun 18;9(6):e0010924. doi: 10.1128/msystems.00109-24. Epub 2024 May 2. mSystems. 2024. PMID: 38695565 Free PMC article. - Chemical genetic approaches for the discovery of bacterial cell wall inhibitors.
Gupta R, Singh M, Pathania R. Gupta R, et al. RSC Med Chem. 2023 Aug 30;14(11):2125-2154. doi: 10.1039/d3md00143a. eCollection 2023 Nov 15. RSC Med Chem. 2023. PMID: 37974958 Free PMC article. Review. - Unrealized targets in the discovery of antibiotics for Gram-negative bacterial infections.
Theuretzbacher U, Blasco B, Duffey M, Piddock LJV. Theuretzbacher U, et al. Nat Rev Drug Discov. 2023 Dec;22(12):957-975. doi: 10.1038/s41573-023-00791-6. Epub 2023 Oct 13. Nat Rev Drug Discov. 2023. PMID: 37833553 Review. - Development, validation, and implementation of an ultratrace analysis method for the determination of moenomycin A, in aquatic animal products.
Tang Y, Yang G, Ma Y, Huang D, Zhai W, Fodjo EK, Zhang X, Li S, Zhang W, Shi Y, Kong C. Tang Y, et al. Anal Bioanal Chem. 2024 Jan;416(3):745-757. doi: 10.1007/s00216-023-04965-4. Epub 2023 Oct 9. Anal Bioanal Chem. 2024. PMID: 37812219
References
- Wallhausser KH, Nesemann G, Prave P, Steigler A. Antimicrob. Agents Chemother. 1965;5:734. A. - PubMed
- Huber G, Schacht U, Weidenmuller HL, Schmidt-Thome J, Duphorn J, Tschesche R. Antimicrob. Agents Chemother. 1965;5:737. - PubMed
- Lenoir D, Tschesche R, Wucherpfennig W, Huber G, Weidenmuller HL. Antimicrob. Agents Chemother. 1969;9:144. - PubMed
- Welzel P. Chem. Rev. 2005;105:4610. - PubMed
- Uchida R, Iwatsuki M, Kim Y-P, Omura S, Tomoda H. J. Antibiot. 2010;63:157. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical