The structure of (CENP-A-H4)(2) reveals physical features that mark centromeres - PubMed (original) (raw)
. 2010 Sep 16;467(7313):347-51.
doi: 10.1038/nature09323. Epub 2010 Aug 25.
Affiliations
- PMID: 20739937
- PMCID: PMC2946842
- DOI: 10.1038/nature09323
The structure of (CENP-A-H4)(2) reveals physical features that mark centromeres
Nikolina Sekulic et al. Nature. 2010.
Abstract
Centromeres are specified epigenetically, and the histone H3 variant CENP-A is assembled into the chromatin of all active centromeres. Divergence from H3 raises the possibility that CENP-A generates unique chromatin features to mark physically centromere location. Here we report the crystal structure of a subnucleosomal heterotetramer, human (CENP-A-H4)(2), that reveals three distinguishing properties encoded by the residues that comprise the CENP-A targeting domain (CATD; ref. 2): (1) a CENP-A-CENP-A interface that is substantially rotated relative to the H3-H3 interface; (2) a protruding loop L1 of the opposite charge as that on H3; and (3) strong hydrophobic contacts that rigidify the CENP-A-H4 interface. Residues involved in the CENP-A-CENP-A rotation are required for efficient incorporation into centromeric chromatin, indicating specificity for an unconventional nucleosome shape. DNA topological analysis indicates that CENP-A-containing nucleosomes are octameric with conventional left-handed DNA wrapping, in contrast to other recent proposals. Our results indicate that CENP-A marks centromere location by restructuring the nucleosome from within its folded histone core.
Figures
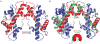
Figure 1. Crystal structure of the (CENP-A/H4)2 heterotetramer
a, Ribbon diagram of the (CENP-A/H4)2 heterotetramer. The structure has clear electron density for residues 59-134 of CENP-A and 24-92 of H4. b, Overlay of (CENP-A/H4)2 with (H3/H4)2 (PDB ID 1KX515) was done with a secondary structure mapping (SSM) algorithm performed with one CENP-A molecule and one H3 molecule from each complex. CENP-A and H3 molecules on the right are overlaid. Similar results are obtained if one molecule of H4 from each complex is used for SSM (rmsd=~0.9 Å in both cases).
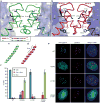
Figure 2. The residues involved in the rotated CENP-A/CENP-A interface are essential for centromere targeting
Detail of H3/H3, a, and CENP-A/CENP-A, b, interfaces with side chains of 5 non-conserved residues in α2 helix at the interface. An alignment of residues in the C-terminal portion of the α2 helix are shown in the box at lower left in panel a. c and d, the α2 helices of H3, c, and CENP-A, d. e, Centromere targeting of WT and mutant versions of CENP-A. f, Quantitation of targeting experiment. For each version of CENP-A, 3 or more experiments were conducted in which a total of >250 cells were analyzed for localization (Cen = exclusively centromeric; Cen + Nuc = both centromere and nucleoplasm staining; Nuc = nucleoplasm only [i.e. no enrichment at centromeres]). Values are plotted +/- s.d.
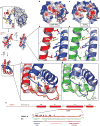
Figure 3. Surface and internal structural features unique to CENP-A-containing complexes
a, The rates of H/DX remapped from the original data are plotted onto the crystal structure of the (CENP-A/H4)2 heterotetramer (left dimer half is shown as ribbon and right dimer half is in surface representation). b and c, Calculated electrostatic potential on the surface of (CENP-A/H4)2, b, and (H3/H4)2, c. The region corresponding to the positively charged bulge in the L1 of CENP-A is circled in each structure. d and e, Hydrophobic interactions between the α2 helices of CENP-A (red) and H4 (blue), d, and α2 helices of H3 (green) and H4 (light blue) e. f and g, Hydrophobic interactions at the helical bundle surrounding the interface of L1 of CENP-A and L2 of H4, f, and L1 of H3 and L2 of H4, g. h, Diagram of the structural differences between CENP-A and H3. Black circles indicate surface exposed residues. The yellow star highlights Arg83 from H3 that inserts into the minor groove of nucleosomal DNA.
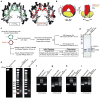
Figure 4. The (CENP-A/H4)2 heterotetramer assembles with H2A/H2B dimers into an octameric nucleosome with conventional handedness of DNA wrapping
a, Basic residues that form contacts with nucleosomal DNA are highlighted on (H3/H4)2 (left) and the putative DNA binding surface of (CENP-A/H4)2 (right). b, Model highlighting alterations in nucleosome structure based on the structure of (CENP-A/H4)2. c, Scheme for nucleosome assembly and analysis. d, Histone content of assembled H3- and CENP-A-containing nucleosomes. e, Digestion of nucleosome arrays with micrococcal nuclease reveals that both H3- and CENP-A-containing nucleosomes protect ~150 bp of DNA. f-i, Topological analysis of H3- containing (f and g) and CENP-A-containing (h and i) nucleosomes. Analysis by gel electrophoresis in the absence (f and h) or presence (g and i) of chloroquine reveals that both H3-containing and CENP-A-containing nucleosomes wrap DNA in a left-handed manner.
Similar articles
- Crystal structure of the human centromeric nucleosome containing CENP-A.
Tachiwana H, Kagawa W, Shiga T, Osakabe A, Miya Y, Saito K, Hayashi-Takanaka Y, Oda T, Sato M, Park SY, Kimura H, Kurumizaka H. Tachiwana H, et al. Nature. 2011 Jul 10;476(7359):232-5. doi: 10.1038/nature10258. Nature. 2011. PMID: 21743476 - Structural determinants for generating centromeric chromatin.
Black BE, Foltz DR, Chakravarthy S, Luger K, Woods VL Jr, Cleveland DW. Black BE, et al. Nature. 2004 Jul 29;430(6999):578-82. doi: 10.1038/nature02766. Nature. 2004. PMID: 15282608 - Comparison between the CENP-A and histone H3 structures in nucleosomes.
Tachiwana H, Kagawa W, Kurumizaka H. Tachiwana H, et al. Nucleus. 2012 Jan-Feb;3(1):6-11. doi: 10.4161/nucl.18372. Nucleus. 2012. PMID: 22127263 - Structure of the CENP-A nucleosome and its implications for centromeric chromatin architecture.
Tachiwana H, Kurumizaka H. Tachiwana H, et al. Genes Genet Syst. 2011;86(6):357-64. doi: 10.1266/ggs.86.357. Genes Genet Syst. 2011. PMID: 22451475 Review. - The histone variant CENP-A and centromere specification.
Black BE, Bassett EA. Black BE, et al. Curr Opin Cell Biol. 2008 Feb;20(1):91-100. doi: 10.1016/j.ceb.2007.11.007. Epub 2008 Jan 15. Curr Opin Cell Biol. 2008. PMID: 18226513 Review.
Cited by
- CENP-A K124 Ubiquitylation Is Required for CENP-A Deposition at the Centromere.
Niikura Y, Kitagawa R, Ogi H, Abdulle R, Pagala V, Kitagawa K. Niikura Y, et al. Dev Cell. 2015 Mar 9;32(5):589-603. doi: 10.1016/j.devcel.2015.01.024. Epub 2015 Feb 26. Dev Cell. 2015. PMID: 25727006 Free PMC article. - Histone variants: emerging players in cancer biology.
Vardabasso C, Hasson D, Ratnakumar K, Chung CY, Duarte LF, Bernstein E. Vardabasso C, et al. Cell Mol Life Sci. 2014 Feb;71(3):379-404. doi: 10.1007/s00018-013-1343-z. Epub 2013 May 8. Cell Mol Life Sci. 2014. PMID: 23652611 Free PMC article. Review. - Plasticity and epigenetic inheritance of centromere-specific histone H3 (CENP-A)-containing nucleosome positioning in the fission yeast.
Yao J, Liu X, Sakuno T, Li W, Xi Y, Aravamudhan P, Joglekar A, Li W, Watanabe Y, He X. Yao J, et al. J Biol Chem. 2013 Jun 28;288(26):19184-96. doi: 10.1074/jbc.M113.471276. Epub 2013 May 9. J Biol Chem. 2013. PMID: 23661703 Free PMC article. - Review series: The functions and consequences of force at kinetochores.
Rago F, Cheeseman IM. Rago F, et al. J Cell Biol. 2013 Mar 4;200(5):557-65. doi: 10.1083/jcb.201211113. J Cell Biol. 2013. PMID: 23460675 Free PMC article. Review. - Structure of the Human Core Centromeric Nucleosome Complex.
Allu PK, Dawicki-McKenna JM, Van Eeuwen T, Slavin M, Braitbard M, Xu C, Kalisman N, Murakami K, Black BE. Allu PK, et al. Curr Biol. 2019 Aug 19;29(16):2625-2639.e5. doi: 10.1016/j.cub.2019.06.062. Epub 2019 Jul 25. Curr Biol. 2019. PMID: 31353180 Free PMC article.
References
- Black BE, Bassett EA. The histone variant CENP-A and centromere specification. Curr Opin Cell Biol. 2008;20:91–100. - PubMed
- Black BE, et al. Structural determinants for generating centromeric chromatin. Nature. 2004;430:578–582. - PubMed
- Mizuguchi G, Xiao H, Wisniewski J, Smith MM, Wu C. Nonhistone Scm3 and histones CenH3-H4 assemble the core of centromere-specific nucleosomes. Cell. 2007;129:1153–1164. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 GM082989/GM/NIGMS NIH HHS/United States
- GM82989/GM/NIGMS NIH HHS/United States
- R01 GM082989-02/GM/NIGMS NIH HHS/United States
- R01 GM082989-03/GM/NIGMS NIH HHS/United States
- R01 GM082989-01A1/GM/NIGMS NIH HHS/United States
- GM08275/GM/NIGMS NIH HHS/United States
- T32 GM008275/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases