Molecular characterization of mucosal adherent bacteria and associations with colorectal adenomas - PubMed (original) (raw)
Molecular characterization of mucosal adherent bacteria and associations with colorectal adenomas
Xiang Jun Shen et al. Gut Microbes. 2010 May-Jun.
Abstract
The human large bowel is colonized by complex and diverse bacterial communities. However, the relationship between commensal bowel bacteria and adenomas (colorectal cancer precursors) is unclear. This study aimed to characterize adherent bacteria in normal colon and evaluate differences in community composition associated with colorectal adenomas. We evaluated adherent bacteria in normal colonic mucosa of 21 adenoma and 23 non-adenoma subjects enrolled in a cross sectional study. Terminal restriction fragment length polymorphism, clone sequencing and fluorescent in-situ hybridization analysis of the 16S rRNA genes were used to characterize adherent bacteria. A total of 335 clones were sequenced and processed for phylogenetic and taxonomic analysis. Differences in bacterial composition between cases and controls were evaluated by UniFrac and analysis of similarity matrix. Overall, Firmicutes (62%), Bacteroidetes (26%) and Proteobacteria (11%) were the most dominant phyla. The bacterial composition differed significantly between cases and controls (UniFrac p < 0.001). We observed significantly higher abundance of Proteobacteria (p < 0.05) and lower abundance of Bacteroidetes (p < 0.05) in cases compared to controls. At the genus level, case subjects showed increased abundance of Dorea spp. (p < 0.005), Faecalibacterium spp. (p < 0.05) and lower proportions of Bacteroides spp. (p < 0.03) and Coprococcus spp. (p < 0.05) than controls. Cases had higher bacterial diversity and richness than controls. These findings reveal that alterations in bacterial community composition associated with adenomas may contribute to the etiology of colorectal cancer. Extension of these findings could lead to strategies to manipulate the microbiota to prevent colorectal adenomas and cancer as well as to identify individuals at high risk.
Keywords: cancer; colorectal adenoma; mucosa adherent bacteria.
Figures
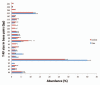
Figure 1
Distribution of T-RFs in adenoma cases and non-adenoma controls. The contribution of each TRF was calculated as a percent of the total TRFs in an individual's sample. Each bar represents the average abundance (y-axis) of each TRF grouped by adenoma case or non-adenoma control status. Error bars represent standard error of the mean. Asterisks-represent TRFs that are significantly different between cases and controls. T-RF, terminal restriction fragment; size in base pairs (bp).
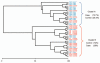
Figure 2
Cluster analysis of T-RF profiles from adenoma cases and non-adenoma controls. Analysis of similarity matrix (ANOSIM) suggests modest differences between the two groups (R = 0.137, p < 0.019). Samples were standardized by calculating each TRF as a percent of the total TRFs in an individual's sample. This was followed by square root transformation and Bray-Curtis similarity matrix was used to generate a dendrogram based on group average. Sample number is presented as H##. Samples from cases and controls are indicated by color code (blue-case; red-control) and those selected for clone library analysis are marked with an asterisk.
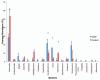
Figure 3A
Distribution of genus level adherent bacterial phylotypes obtained from clone libraries. Each bar represents the percent contribution of genus-level profiles grouped by case-control status (A) or for each individual (B). The colors representing the different genera are shown on the side of the figures.
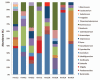
Figure 3B
Distribution of genus level adherent bacterial phylotypes obtained from clone libraries. Each bar represents the percent contribution of genus-level profiles grouped by case-control status (A) or for each individual (B). The colors representing the different genera are shown on the side of the figures.
Figure 4
Fluorescence in-situ hybridization (FISH) using bacterial 16S rRNA probes showing bacteria localized to the mucus layer. (A) H&E stained section of normal colonic mucosa showing crypts and mucosal layer. Tissue sections from colonic mucosa were hybridized with general bacteria (EUB 388) probe: (B and C) show that general bacteria (include most of Eubacteria species) were distributed in the mucosal layer; (B) shows bacteria (red) at magnification, 400X while (C) shows confocal laser scanning image of bacteria (red) in mucosa at a higher magnification. Colon tissue sections were also hybridized with specific bacteria probes namely Faecalibacterium, Proteobacteria, Clostridia, Ruminococcus and Lactobacillus (D–H). (D and E) were stained with bacteria specific probe and general bacteria probe. (F–H) were stained with single bacteria specific probe. Mammalian cell nuclei are stained blue by DAPI staining (D and F). Orange arrows show different shaped bacteria localized to the mucus layer. White arrow points to the mucus layer above the colonic epithelium.
References
- Savage DC. Microbial ecology of the gastrointestinal tract. Ann Rev Microbiol. 1977;31:107–133. - PubMed
- Van Citters GW, Lin HC. Management of small intestinal bacterial overgrowth. Curr Gastroenterol Rep. 2005;7:317–320. - PubMed
- Mutch DM, Simmering R, Donnicola D, Fotopoulos G, Holzwarth JA, Williamson G, et al. Impact of commensal microbiota on murine gastrointestinal tract gene ontologies. Physiol Genomics. 2004;19:22–31. - PubMed
- Hasegawa M, Kawasaki A, Yang K, Fujimoto Y, Masumoto J, Breukink E, et al. A role of lipophilic peptidoglycan-related molecules in induction of Nod1-mediated immune responses. J Biol Chem. 2007;282:11757–11764. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P30 ES010126/ES/NIEHS NIH HHS/United States
- R01 CA044684/CA/NCI NIH HHS/United States
- R01-CA136887/CA/NCI NIH HHS/United States
- R01 DK081426/DK/NIDDK NIH HHS/United States
- R01 CA136887/CA/NCI NIH HHS/United States
- K01 DK073695/DK/NIDDK NIH HHS/United States
- P50 CA106991/CA/NCI NIH HHS/United States
- K01 CA093654/CA/NCI NIH HHS/United States
- P30 DK034987/DK/NIDDK NIH HHS/United States
- P30 DK34987/DK/NIDDK NIH HHS/United States
- R01 CA44684/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous