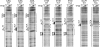The TCT motif, a key component of an RNA polymerase II transcription system for the translational machinery - PubMed (original) (raw)
The TCT motif, a key component of an RNA polymerase II transcription system for the translational machinery
Trevor J Parry et al. Genes Dev. 2010.
Abstract
The TCT motif (polypyrimidine initiator) encompasses the transcription start site of nearly all ribosomal protein genes in Drosophila and mammals. The TCT motif is required for transcription of ribosomal protein gene promoters. The TCT element resembles the Inr (initiator), but is not recognized by TFIID and cannot function in lieu of an Inr. However, a single T-to-A substitution converts the TCT element into a functionally active Inr. Thus, the TCT motif is a novel transcriptional element that is distinct from the Inr. These findings reveal a specialized TCT-based transcription system that is directed toward the synthesis of ribosomal proteins.
Figures
Figure 1.
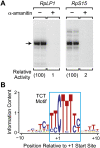
Analysis of core promoters of Drosophila RP genes. (A) Transcription of RP genes by RNA polymerase II. Core promoter constructs containing sequences from −50 to +50 (relative to the C+1 transcription start site) of the RpLP1 and RpS15 genes were subjected to in vitro transcription analysis in the absence or presence of 4 μg/mL α-amanitin. The resulting transcripts were detected by primer extension analysis. (B) DNA sequence logo of the TCT motif, which encompasses the transcription start site region of RP genes. The sequence logo was generated as described elsewhere (Schneider and Stephens 1990; Crooks et al. 2004) with the core promoter sequence data in Supplemental Table 1.
Figure 2.
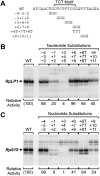
Sequences in the TCT motif are critical for transcription from RP gene core promoters. (A) Diagram of scanning triple-nucleotide substitution mutations. The trinucleotides at the specified positions (relative to the C+1) were converted to GGG, except as indicated for the +6T+7T+8T mutant promoter. The sequence of the Drosophila RpLP1 core promoter is shown. An analogous series of mutations was constructed with the RpS15 core promoter. (B,C) In vitro transcription analyses of the wild-type and mutant RpLP1 and RpS15 promoters. The transcriptional activity of each mutant promoter is reported relative to that of its cognate wild-type promoter.
Figure 3.
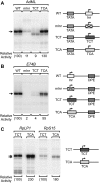
A single T-to-A substitution can convert a TCT element, which does not function with TATA or DPE motifs, into an Inr. (A) The TCT motif does not function with a TATA box in lieu of an Inr. Wild-type and mutant versions of the AdML core promoter were subjected to in vitro transcription analysis. In the mInr promoter, the Inr (TCACTC) was mutated to GTGACA. In the TCT promoter, the AdML Inr and flanking nucleotides (CTCACTCT) were replaced with the corresponding TCT sequence (CTCTTTCC) from the RpLP1 core promoter. The TCA promoter contains a T-to-A substitution in the TCT motif (CTC
A
TTCC). The transcriptional activity of each mutant promoter is reported relative to that of the wild-type AdML promoter. (B) The TCT motif does not function with a DPE in lieu of an Inr. Wild-type and mutant versions of the DPE-dependent E74B core promoter were generated and analyzed as described in A with the AdML promoter. (C) The TCT-to-TCA mutation in RP gene promoters results in a downstream shift of the transcription start site. The TCT sequence in the TCT motifs of the RpLP1 and RpS15 core promoters were mutated to TCA, and the resulting wild-type and mutant constructs were subjected to in vitro transcription analysis.
Figure 4.
A single T-to-A substitution in the TCT motif substantially increases the affinity of TFIID binding to the core promoter. DNase I footprinting analyses were carried out with purified Drosophila TFIID. With the AdML and E74B promoter constructs, the five footprint reactions contained 0, 30, 60, 90, and 0 ng of TFIID. With the RpLP1 promoter constructs, the reactions contained 0, 15, 30, 45, and 0 ng of TFIID. Regions of DNase I protection and hypersensitivity are indicated by brackets and solid dots, respectively.
Similar articles
- The MTE, a new core promoter element for transcription by RNA polymerase II.
Lim CY, Santoso B, Boulay T, Dong E, Ohler U, Kadonaga JT. Lim CY, et al. Genes Dev. 2004 Jul 1;18(13):1606-17. doi: 10.1101/gad.1193404. Genes Dev. 2004. PMID: 15231738 Free PMC article. - CIF, an essential cofactor for TFIID-dependent initiator function.
Kaufmann J, Verrijzer CP, Shao J, Smale ST. Kaufmann J, et al. Genes Dev. 1996 Apr 1;10(7):873-86. doi: 10.1101/gad.10.7.873. Genes Dev. 1996. PMID: 8846923 - Site-specific initiation of transcription by RNA polymerase II.
Kollmar R, Farnham PJ. Kollmar R, et al. Proc Soc Exp Biol Med. 1993 Jun;203(2):127-39. doi: 10.3181/00379727-203-43583. Proc Soc Exp Biol Med. 1993. PMID: 8502653 Review. - TATA-binding protein-associated factor(s) in TFIID function through the initiator to direct basal transcription from a TATA-less class II promoter.
Martinez E, Chiang CM, Ge H, Roeder RG. Martinez E, et al. EMBO J. 1994 Jul 1;13(13):3115-26. doi: 10.1002/j.1460-2075.1994.tb06610.x. EMBO J. 1994. PMID: 7518774 Free PMC article. - Transcription by RNA polymerase II: initiator-directed formation of transcription-competent complexes.
Weis L, Reinberg D. Weis L, et al. FASEB J. 1992 Nov;6(14):3300-9. doi: 10.1096/fasebj.6.14.1426767. FASEB J. 1992. PMID: 1426767 Review.
Cited by
- Functionally distinct promoter classes initiate transcription via different mechanisms reflected in focused versus dispersed initiation patterns.
Serebreni L, Pleyer LM, Haberle V, Hendy O, Vlasova A, Loubiere V, Nemčko F, Bergauer K, Roitinger E, Mechtler K, Stark A. Serebreni L, et al. EMBO J. 2023 May 15;42(10):e113519. doi: 10.15252/embj.2023113519. Epub 2023 Apr 4. EMBO J. 2023. PMID: 37013908 Free PMC article. - Metazoan promoters: emerging characteristics and insights into transcriptional regulation.
Lenhard B, Sandelin A, Carninci P. Lenhard B, et al. Nat Rev Genet. 2012 Mar 6;13(4):233-45. doi: 10.1038/nrg3163. Nat Rev Genet. 2012. PMID: 22392219 Review. - Intra-promoter switch of transcription initiation sites in proliferation signaling-dependent RNA metabolism.
Wragg JW, White PL, Hadzhiev Y, Wanigasooriya K, Stodolna A, Tee L, Barros-Silva JD, Beggs AD, Müller F. Wragg JW, et al. Nat Struct Mol Biol. 2023 Dec;30(12):1970-1984. doi: 10.1038/s41594-023-01156-8. Epub 2023 Nov 23. Nat Struct Mol Biol. 2023. PMID: 37996663 Free PMC article. - Identifying unique exposure-specific transgenerational differentially DNA methylated region epimutations in the genome using hybrid deep learning prediction models.
Mavaie P, Holder L, Skinner M. Mavaie P, et al. Environ Epigenet. 2023 Nov 30;9(1):dvad007. doi: 10.1093/eep/dvad007. eCollection 2023. Environ Epigenet. 2023. PMID: 38130880 Free PMC article. - Delineation of the DNA Structural Features of Eukaryotic Core Promoter Classes.
Vanaja A, Yella VR. Vanaja A, et al. ACS Omega. 2022 Feb 9;7(7):5657-5669. doi: 10.1021/acsomega.1c04603. eCollection 2022 Feb 22. ACS Omega. 2022. PMID: 35224327 Free PMC article.
References
- Burke TW, Kadonaga JT 1996. Drosophila TFIID binds to a conserved downstream basal promoter element that is present in many TATA-box-deficient promoters. Genes Dev 10: 711–724 - PubMed
- Chung S, Perry RP 1991. Cell-free transcription of a mouse ribosomal-protein-encoding gene: The effects of promoter mutations. Gene 100: 173–180 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases