Spotlight on post-transcriptional control in the circadian system - PubMed (original) (raw)
Review
Spotlight on post-transcriptional control in the circadian system
Dorothee Staiger et al. Cell Mol Life Sci. 2011 Jan.
Abstract
An endogenous timing mechanism, the circadian clock, causes rhythmic expression of a considerable fraction of the genome of most organisms to optimally align physiology and behavior with their environment. Circadian clocks are self-sustained oscillators primarily based on transcriptional feedback loops and post-translational modification of clock proteins. It is increasingly becoming clear that regulation at the RNA level strongly impacts the cellular circadian transcriptome and proteome as well as the oscillator mechanism itself. This review focuses on posttranscriptional events, discussing RNA-binding proteins that, by influencing the timing of pre-mRNA splicing, polyadenylation and RNA decay, shape rhythmic expression profiles. Furthermore, recent findings on the contribution of microRNAs to orchestrating circadian rhythms are summarized.
Figures
Fig. 1
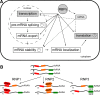
Post-transcriptional regulation of gene expression. a The steps of pre-mRNA processing controlled by RNA-binding proteins or microRNAs. Clock symbols denote events known to be associated with circadian regulation. b Post-transcriptional operons defined by RBPs or miRNAs binding to cognate cis-regulatory elements in mRNAs [11]
Fig. 2
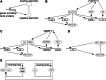
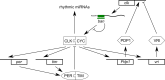
Transcriptional clock feedback loops. Common design principle (a) and model of the Drosophila (b), mammalian (c) and Neurospora (d) oscillator. See text for details. e Simplified scheme of the Arabidopsis oscillator consisting of the central CCA1/LHY-TOC1 loop, a morning-phase loop and an evening-phased loop [2, 32]
Fig. 3
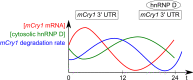
Scheme of the inverse mCry1 mRNA and cytoplasmic hnRNP D profiles. Relative levels of mCry1, hnRNP D protein abundance in the cytoplasm and mCry1 degradation rate across 1 day are shown (adapted from data in [43])
Fig. 4
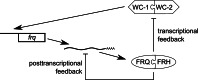
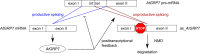
Transcriptional and post-transcriptional regulation by FRH. In the transcriptional feedback loop, FRQ and FRH inhibit WCC activity leading to repression of frq transcription. In the post-transcriptional feedback loop, FRQ and FRH promote frq decay through the exosome
Fig. 5
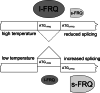
Temperature-dependent alternative splicing of an intron encompassing the l-FRQ start codon regulates the ratio of l-FRQ versus s-FRQ. The sizes of l-FRQ and s-FRQ symbols reflect their relative amounts
Fig. 6
Negative autoregulation of the RNA-binding protein _At_GRP7 through alternative splicing and subsequent nonsense-mediated decay (NMD) [70]. In the presence of elevated _At_GRP7 protein levels a higher proportion of AtGRP7 pre-mRNA is spliced to a PTC-containing NMD substrate through binding of _At_GRP7 to its own intron and subsequent use of a cryptic intronic 5′splice site. The alternative splice form (as_ AtGRP7) contains a PTC and is degraded via NMD
Fig. 7
Involvement of the miRNAs in the Drosophila circadian system. The core oscillator controls rhythmic expression of a number of miRNAs [93]. The bantam miRNA targets the oscillator component CLK through interaction with its 3′UTR [94]
Similar articles
- RNA-based regulation in the plant circadian clock.
Staiger D, Green R. Staiger D, et al. Trends Plant Sci. 2011 Oct;16(10):517-23. doi: 10.1016/j.tplants.2011.06.002. Epub 2011 Jul 23. Trends Plant Sci. 2011. PMID: 21782493 Review. - Post-transcriptional control of circadian rhythms.
Kojima S, Shingle DL, Green CB. Kojima S, et al. J Cell Sci. 2011 Feb 1;124(Pt 3):311-20. doi: 10.1242/jcs.065771. J Cell Sci. 2011. PMID: 21242310 Free PMC article. Review. - Post-transcriptional modulators and mediators of the circadian clock.
Anna G, Kannan NN. Anna G, et al. Chronobiol Int. 2021 Sep;38(9):1244-1261. doi: 10.1080/07420528.2021.1928159. Epub 2021 May 31. Chronobiol Int. 2021. PMID: 34056966 Free PMC article. Review. - Adaptation of molecular circadian clockwork to environmental changes: a role for alternative splicing and miRNAs.
Bartok O, Kyriacou CP, Levine J, Sehgal A, Kadener S. Bartok O, et al. Proc Biol Sci. 2013 Jul 3;280(1765):20130011. doi: 10.1098/rspb.2013.0011. Print 2013 Aug 22. Proc Biol Sci. 2013. PMID: 23825200 Free PMC article. Review. - Circadian clocks and natural antisense RNA.
Crosthwaite SK. Crosthwaite SK. FEBS Lett. 2004 Jun 1;567(1):49-54. doi: 10.1016/j.febslet.2004.04.073. FEBS Lett. 2004. PMID: 15165892 Review.
Cited by
- RNA around the clock - regulation at the RNA level in biological timing.
Nolte C, Staiger D. Nolte C, et al. Front Plant Sci. 2015 May 5;6:311. doi: 10.3389/fpls.2015.00311. eCollection 2015. Front Plant Sci. 2015. PMID: 25999975 Free PMC article. Review. - An Untranslated cis-Element Regulates the Accumulation of Multiple C4 Enzymes in Gynandropsis gynandra Mesophyll Cells.
Williams BP, Burgess SJ, Reyna-Llorens I, Knerova J, Aubry S, Stanley S, Hibberd JM. Williams BP, et al. Plant Cell. 2016 Feb;28(2):454-65. doi: 10.1105/tpc.15.00570. Epub 2016 Jan 15. Plant Cell. 2016. PMID: 26772995 Free PMC article. - Comparative Proteomic Analysis of the Response of Maize (Zea mays L.) Leaves to Long Photoperiod Condition.
Wu L, Tian L, Wang S, Zhang J, Liu P, Tian Z, Zhang H, Liu H, Chen Y. Wu L, et al. Front Plant Sci. 2016 Jun 2;7:752. doi: 10.3389/fpls.2016.00752. eCollection 2016. Front Plant Sci. 2016. PMID: 27313588 Free PMC article. - Circadian Rhythm and Sleep Disruption: Causes, Metabolic Consequences, and Countermeasures.
Potter GD, Skene DJ, Arendt J, Cade JE, Grant PJ, Hardie LJ. Potter GD, et al. Endocr Rev. 2016 Dec;37(6):584-608. doi: 10.1210/er.2016-1083. Epub 2016 Oct 20. Endocr Rev. 2016. PMID: 27763782 Free PMC article. Review. - Diurnal rhythms in neurexins transcripts and inhibitory/excitatory synapse scaffold proteins in the biological clock.
Shapiro-Reznik M, Jilg A, Lerner H, Earnest DJ, Zisapel N. Shapiro-Reznik M, et al. PLoS One. 2012;7(5):e37894. doi: 10.1371/journal.pone.0037894. Epub 2012 May 25. PLoS One. 2012. PMID: 22662246 Free PMC article.
References
- Gachon F, Nagoshi E, Brown SA, Ripperger J, Schibler U. The mammalian circadian timing system: from gene expression to physiology. Chromosoma. 2004;113:103–112. - PubMed
- Mas P. Circadian clock function in Arabidopsis thaliana: time beyond transcription. Trends Cell Biol. 2008;18:273–281. - PubMed
- Schöning JC, Staiger D. At the pulse of time: protein interactions determine the pace of circadian clocks. FEBS Lett. 2005;579:3246–3252. - PubMed
- Gallego M, Virshup DM. Post-translational modifications regulate the ticking of the circadian clock. Nat Rev Mol Cell Biol. 2007;8:139–148. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases