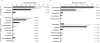Gut microbial gene expression in mother-fed and formula-fed piglets - PubMed (original) (raw)
Gut microbial gene expression in mother-fed and formula-fed piglets
Valeriy Poroyko et al. PLoS One. 2010.
Abstract
Background: Effects of diet on the structure and function of gut microbial communities in newborn infants are poorly understood. High-resolution molecular studies are needed to definitively ascertain whether gut microbial communities are distinct in milk-fed and formula-fed infants.
Methodology/principal findings: Pyrosequencing-based whole transcriptome shotgun sequencing (RNA-seq) was used to evaluate community wide gut microbial gene expression in 21 day old neonatal piglets fed either with sow's milk (mother fed, MF; n = 4) or with artificial formula (formula fed, FF; n = 4). Microbial DNA and RNA were harvested from cecal contents for each animal. cDNA libraries and 16S rDNA amplicons were sequenced on the Roche 454 GS-FLX Titanium system. Communities were similar at the level of phylum but were dissimilar at the level of genus; Prevotella was the dominant genus within MF samples and Bacteroides was most abundant within FF samples. Screened cDNA sequences were assigned functional annotations by the MG-RAST annotation pipeline and based upon best-BLASTX-hits to the NCBI COG database. Patterns of gene expression were very similar in MF and FF animals. All samples were enriched with transcripts encoding enzymes for carbohydrate and protein metabolism, as well as proteins involved in stress response, binding to host epithelium, and lipopolysaccharide metabolism. Carbohydrate utilization transcripts were generally similar in both groups. The abundance of enzymes involved in several pathways related to amino acid metabolism (e.g., arginine metabolism) and oxidative stress response differed in MF and FF animals.
Conclusions/significance: Abundant transcripts identified in this study likely contribute to a core microbial metatranscriptome in the distal intestine. Although microbial community gene expression was generally similar in the cecal contents of MF and FF neonatal piglets, several differentially abundant gene clusters were identified. Further investigations of gut microbial gene expression will contribute to a better understanding of normal and abnormal enteric microbiology in animals and humans.
Conflict of interest statement
Competing Interests: SD has received research funding from Bristol Myers Squibb. This company played no role in the planning, conduct, or analysis of the experiments in this manuscript. This relationship does not alter the authors' adherence to PLoS ONE policies on sharing data and materials.
Figures
Figure 1. Microbial community structure in cecal contents from mother-fed and formula-fed piglets at 21 days of life.
(A) Mean relative abundances of bacterial taxa within cecal microbiota from 4 MF animals (black bars) and 4 FF animals (white bars) identified by analysis of 16S rDNA amplicon sequences. Sequences were classified to the highest taxonomic level to which they could be confidently assigned using the RDP classification algorithm and taxonomic hierarchy. (B) Relative mean abundances of bacterial taxa within cecal microbiota identified by RDP analysis of unamplified 16S sequences within cDNA libraries. The results demonstrate that all communities are dominated by the phyla Bacteroidetes and Firmicutes; MF data sets are enriched with Prevotella sequences and FF data sets are enriched with Bacteroides sequences. In both panels, differentially abundant phyla (p value<0.05) and genera (p value<0.01) are marked with a * if they were enriched within the MF samples and with a ** if they were enriched within FF samples. Taxa with a mean relative abundance <0.01 in both MF and FF groups are not shown.
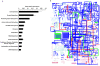
Figure 2. A core microbial metatranscriptome in the piglet cecum.
(A) Mean relative abundances of annotated sequences within cDNA libraries from all 8 animals studied. Displayed are the automated SEED Level 1 Subsystem assignments, as determined by MG-RAST . Low standard deviations indicate that variation in the gut metatranscriptome between subjects is low. (B) Projection of the global metabolic profiles onto the KEGG pathways using the iPath tool demonstrates the overall similarity of the MF and FF gut microbial communities. Metabolic pathways common to both diets are shown in blue. Pathways unique to the MF animals are represented in green, and pathways unique to the FF animals are represented in red.
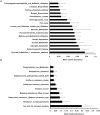
Figure 3. Universal expression of carbohydrate utilization and microbial virulence genes in the piglet cecum.
(A) Relative abundance of cDNA sequences assigned by MG-RAST to the Level 3 SEED Subsystem of carbohydrate utilization. (B) Relative abundance of cDNA sequences assigned by MG-RAST to the Level 3 SEED Subsystem of virulence. Values for mean relative abundances in both (A) and (B) reflect average values across all 8 animals studied.
Figure 4. Relationship between gut microbial community structure and function.
(A) Mean relative abundances of bacterial taxa within cecal microbiota from 4 MF animals (black bars) and 4 FF animals (white bars) identified by analysis of non-ribosomal sequences within cDNA libraries. Taxonomic assignments for sequenced transcripts were made by identifying best BLASTN hits against an in-house database of microbial genomes. Differentially abundant phyla (p value<0.05) and genera (p value<0.01) are marked with a * if they were enriched within the MF samples and with a ** if they were enriched within FF samples. (B) Taxonomic origin of highly expressed genes. Expression levels for the two most abundant general COG hits and two most abundant individual COG hits are represented for each of the four most abundant bacterial phyla.
Similar articles
- Murine gut microbiota and transcriptome are diet dependent.
Carlisle EM, Poroyko V, Caplan MS, Alverdy J, Morowitz MJ, Liu D. Carlisle EM, et al. Ann Surg. 2013 Feb;257(2):287-94. doi: 10.1097/SLA.0b013e318262a6a6. Ann Surg. 2013. PMID: 23001074 - Bacterial and Fungal Adaptations in Cecum and Distal Colon of Piglets Fed With Dairy-Based Milk Formula in Comparison With Human Milk.
Elolimy A, Rosa F, Tripp P, Zeineldin M, Bowlin AK, Randolph C, Robeson MS, Yeruva L. Elolimy A, et al. Front Microbiol. 2022 Mar 23;13:801854. doi: 10.3389/fmicb.2022.801854. eCollection 2022. Front Microbiol. 2022. PMID: 35401465 Free PMC article. - Milk Formula Diet Alters Bacterial and Host Protein Profile in Comparison to Human Milk Diet in Neonatal Piglet Model.
Rosa F, Zybailov BL, Glazko GV, Rahmatallah Y, Byrum S, Mackintosh SG, Bowlin AK, Yeruva L. Rosa F, et al. Nutrients. 2021 Oct 22;13(11):3718. doi: 10.3390/nu13113718. Nutrients. 2021. PMID: 34835974 Free PMC article. - Neonatal Diet Impacts Bioregional Microbiota Composition in Piglets Fed Human Breast Milk or Infant Formula.
Brink LR, Matazel K, Piccolo BD, Bowlin AK, Chintapalli SV, Shankar K, Yeruva L. Brink LR, et al. J Nutr. 2019 Dec 1;149(12):2236-2246. doi: 10.1093/jn/nxz170. J Nutr. 2019. PMID: 31373372 Free PMC article. - Host-microbe interactions in the neonatal intestine: role of human milk oligosaccharides.
Donovan SM, Wang M, Li M, Friedberg I, Schwartz SL, Chapkin RS. Donovan SM, et al. Adv Nutr. 2012 May 1;3(3):450S-5S. doi: 10.3945/an.112.001859. Adv Nutr. 2012. PMID: 22585924 Free PMC article. Review.
Cited by
- Functional similarities between pigeon 'milk' and mammalian milk: induction of immune gene expression and modification of the microbiota.
Gillespie MJ, Stanley D, Chen H, Donald JA, Nicholas KR, Moore RJ, Crowley TM. Gillespie MJ, et al. PLoS One. 2012;7(10):e48363. doi: 10.1371/journal.pone.0048363. Epub 2012 Oct 26. PLoS One. 2012. PMID: 23110233 Free PMC article. - Alterations in the porcine colon microbiota induced by the gastrointestinal nematode Trichuris suis.
Li RW, Wu S, Li W, Navarro K, Couch RD, Hill D, Urban JF Jr. Li RW, et al. Infect Immun. 2012 Jun;80(6):2150-7. doi: 10.1128/IAI.00141-12. Epub 2012 Apr 9. Infect Immun. 2012. PMID: 22493085 Free PMC article. - Analysis of muscle and ovary transcriptome of Sus scrofa: assembly, annotation and marker discovery.
Nie Q, Fang M, Jia X, Zhang W, Zhou X, He X, Zhang X. Nie Q, et al. DNA Res. 2011 Oct;18(5):343-51. doi: 10.1093/dnares/dsr021. Epub 2011 Jul 5. DNA Res. 2011. PMID: 21729922 Free PMC article. - Changes of gut microbiota structure and morphology in weaned piglets treated with fresh fermented soybean meal.
Xie Z, Hu L, Li Y, Geng S, Cheng S, Fu X, Zhao S, Han X. Xie Z, et al. World J Microbiol Biotechnol. 2017 Nov 16;33(12):213. doi: 10.1007/s11274-017-2374-7. World J Microbiol Biotechnol. 2017. PMID: 29147865 - High-throughput sequence-based analysis of the intestinal microbiota of weanling pigs fed genetically modified MON810 maize expressing Bacillus thuringiensis Cry1Ab (Bt maize) for 31 days.
Buzoianu SG, Walsh MC, Rea MC, O'Sullivan O, Cotter PD, Ross RP, Gardiner GE, Lawlor PG. Buzoianu SG, et al. Appl Environ Microbiol. 2012 Jun;78(12):4217-24. doi: 10.1128/AEM.00307-12. Epub 2012 Mar 30. Appl Environ Microbiol. 2012. PMID: 22467509 Free PMC article.
References
- Mackie RI, Sghir A, Gaskins HR. Developmental microbial ecology of the neonatal gastrointestinal tract. Am J Clin Nutr. 1999;69:1035S–45S. - PubMed
- Penders J, Thijs C, Vink C, Stelma FF, Snijders B, et al. Factors influencing the composition of the intestinal microbiota in early infancy. Pediatrics. 2006;118:511–21. - PubMed
- Lundequist B, Nord CE, Winberg J. The composition of the faecal microflora in breastfed and bottle fed infants from birth to eight weeks. Acta Paediatr Scand 1985. 1985;74:45–51. - PubMed
- Hopkins MJ, Macfarlane GT, Furrie E, Fite A, Macfarlane S. Characterisation of intestinal bacteria in infant stools using real-time PCR and northern hybridisation analyses. FEMS Microbiol Ecol. 2005;54:77–85. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources