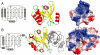Identification and characterization of two families of F420 H2-dependent reductases from Mycobacteria that catalyse aflatoxin degradation - PubMed (original) (raw)
doi: 10.1111/j.1365-2958.2010.07356.x. Epub 2010 Sep 16.
Colin J Jackson, David B Tattersall, Nigel French, Thomas S Peat, Janet Newman, Lyndall J Briggs, Gauri V Lapalikar, Peter M Campbell, Colin Scott, Robyn J Russell, John G Oakeshott
Affiliations
- PMID: 20807200
- PMCID: PMC3034190
- DOI: 10.1111/j.1365-2958.2010.07356.x
Free PMC article
Identification and characterization of two families of F420 H2-dependent reductases from Mycobacteria that catalyse aflatoxin degradation
Matthew C Taylor et al. Mol Microbiol. 2010 Nov.
Free PMC article
Abstract
Aflatoxins are polyaromatic mycotoxins that contaminate a range of food crops as a result of fungal growth and contribute to serious health problems in the developing world because of their toxicity and mutagenicity. Although relatively resistant to biotic degradation, aflatoxins can be metabolized by certain species of Actinomycetales. However, the enzymatic basis for their breakdown has not been reported until now. We have identified nine Mycobacterium smegmatis enzymes that utilize the deazaflavin cofactor F(420) H(2) to catalyse the reduction of the α,β-unsaturated ester moiety of aflatoxins, activating the molecules for spontaneous hydrolysis and detoxification. These enzymes belong to two previously uncharacterized F(420) H(2) dependent reductase (FDR-A and -B) families that are distantly related to the flavin mononucleotide (FMN) dependent pyridoxamine 5'-phosphate oxidases (PNPOxs). We have solved crystal structures of an enzyme from each FDR family and show that they, like the PNPOxs, adopt a split barrel protein fold, although the FDRs also possess an extended and highly charged F(420) H(2) binding groove. A general role for these enzymes in xenobiotic metabolism is discussed, including the observation that the nitro-reductase Rv3547 from Mycobacterium tuberculosis that is responsible for the activation of bicyclic nitroimidazole prodrugs belongs to the FDR-A family.
© 2010 Blackwell Publishing Ltd.
Figures
Fig. 1
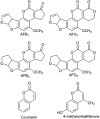
Chemical structures of compounds used in this study.
Fig. 2
Purification protocol. Protein fractions that showed F420H2 dependent AFG1 degradation as measured by TLC were separated from M. smegmatis extracts. The ammonium sulphate precipitated proteins were first purified by hydrophobic interaction chromatography (HIC). Active fractions were further purified by anion exchange chromatography before separation by SDS-PAGE. Bands were cut from SDS-PAGE gels, digested with trypsin and analysed by LC/MS/MS. Peptides were identified from the annotated M. smegmatis genome sequence and corresponding results are shown for some of the excised bands. In the TLCs ‘-ve’ denotes no enzyme negative control and ‘sm’ denotes M. smegmatis cell extract positive control.
Fig. 3
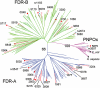
Phylogenetic relationship between the PNPOx, FDR-A and FDR-B families. A condensed tree was constructed from 146 protein sequences of FDRs/PNPOxs from seven species of Actinomycetales (M. smegmatis, M. tuberculosis H37Rv, M. vanbaalenii, Rhodococcus sp. RH1, Arthrobacter sp. FB24, S. coelicolor, Frankia alni and Nocardioides sp. JS614), plus the known PNPOx enzymes from H. sapiens, E. coli, Saccharomyces cerevisiae, Caenorhabditis elegans and Mus musculus. Solid red circles denote the FDRs described in this paper, and open red circles denote other potential FDRs from M. smegmatis (TIGR locus number shown). For clarity, only enzymes from other species that have been previously characterized are labelled: blue triangles denote M. tuberculosis enzymes with known structures or previously described functions and black squares denote previously described PNPOxs.
Fig. 4
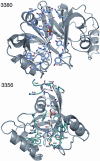
The structures of FDR-A and FDR-B enzymes. A. MSMEG_3356; from left to right, a topology diagram, showing the arrangement of the split barrel of anti-parallel β-sheets, surrounded by three large α-helices at the side and a short α-helix above, a cartoon diagram, and an electrostatic surface potential representation with F420H2 docked. B. MSMEG_3380; from left to right, a topology diagram of the structure, showing the arrangement of the split barrel of anti-parallel β-sheets, surrounded by one large α-helix at the side and three short α-helices above and below, a cartoon diagram of the 3380 dimer, and an electrostatic surface potential representation with F420H2 docked.
Fig. 5
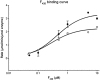
F420H2 cofactor binding to enzymes 3356 and 3380. The apparent _K_M's for F420H2 were calculated by measuring the velocity of the reaction with varying concentrations of F420H2 from 0.05 to 10 µM while maintaining all other components at 100 µM aflatoxin, 1 µM enzyme. For clarity the velocity is expressed as percent of maximum velocity. Empty circles, 3356; filled circles, 3380.
Fig. 6
The F420H2 binding grooves of an FDR-A, MSMEG_3356, and an FDR-B, MSMEG_3380. The deazaflavin pockets are shown to be rich in aromatic and hydrophobic residues, while the side-chain binding grooves are rich in basic residues.
Fig. 7
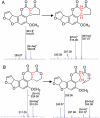
Proposed reaction mechanism of AFB1 and AFG1 reduction by reduced F420. A. The proposed reduction mechanism of AFB1, with the mass spectrum of the substrate and product below the chemical structures. B. The mass spectrum and chemical structure of AFG1 and the reduced product.
Similar articles
- A new role for coenzyme F420 in aflatoxin reduction by soil mycobacteria.
Graham DE. Graham DE. Mol Microbiol. 2010 Nov;78(3):533-6. doi: 10.1111/j.1365-2958.2010.07358.x. Mol Microbiol. 2010. PMID: 21038477 - Sequence-Structure-Function Classification of a Catalytically Diverse Oxidoreductase Superfamily in Mycobacteria.
Ahmed FH, Carr PD, Lee BM, Afriat-Jurnou L, Mohamed AE, Hong NS, Flanagan J, Taylor MC, Greening C, Jackson CJ. Ahmed FH, et al. J Mol Biol. 2015 Nov 6;427(22):3554-3571. doi: 10.1016/j.jmb.2015.09.021. Epub 2015 Oct 3. J Mol Biol. 2015. PMID: 26434506 - F420H2-dependent degradation of aflatoxin and other furanocoumarins is widespread throughout the actinomycetales.
Lapalikar GV, Taylor MC, Warden AC, Scott C, Russell RJ, Oakeshott JG. Lapalikar GV, et al. PLoS One. 2012;7(2):e30114. doi: 10.1371/journal.pone.0030114. Epub 2012 Feb 27. PLoS One. 2012. PMID: 22383957 Free PMC article. - Physiology, Biochemistry, and Applications of F420- and Fo-Dependent Redox Reactions.
Greening C, Ahmed FH, Mohamed AE, Lee BM, Pandey G, Warden AC, Scott C, Oakeshott JG, Taylor MC, Jackson CJ. Greening C, et al. Microbiol Mol Biol Rev. 2016 Apr 27;80(2):451-93. doi: 10.1128/MMBR.00070-15. Print 2016 Jun. Microbiol Mol Biol Rev. 2016. PMID: 27122598 Free PMC article. Review. - Molybdenum enzymes and molybdenum cofactor in mycobacteria.
Shi T, Xie J. Shi T, et al. J Cell Biochem. 2011 Oct;112(10):2721-8. doi: 10.1002/jcb.23233. J Cell Biochem. 2011. PMID: 21678480 Review.
Cited by
- Mechanisms by which microbial enzymes degrade four mycotoxins and application in animal production: A review.
Sun H, He Z, Xiong D, Long M. Sun H, et al. Anim Nutr. 2023 Oct 4;15:256-274. doi: 10.1016/j.aninu.2023.09.003. eCollection 2023 Dec. Anim Nutr. 2023. PMID: 38033608 Free PMC article. Review. - Functional Characterization and Whole-Genome Analysis of an Aflatoxin-Degrading Rhodococcus pyridinivorans Strain.
Deng D, Tang J, Liu Z, Tian Z, Song M, Cui Y, Rong T, Lu H, Yu M, Li J, Pang R, Ma X. Deng D, et al. Biology (Basel). 2022 May 19;11(5):774. doi: 10.3390/biology11050774. Biology (Basel). 2022. PMID: 35625502 Free PMC article. - Current and Emerging Tools of Computational Biology To Improve the Detoxification of Mycotoxins.
Sandlin N, Russell Kish D, Kim J, Zaccaria M, Momeni B. Sandlin N, et al. Appl Environ Microbiol. 2022 Feb 8;88(3):e0210221. doi: 10.1128/AEM.02102-21. Epub 2021 Dec 8. Appl Environ Microbiol. 2022. PMID: 34878810 Free PMC article. - Detoxification of aflatoxin B1 by a Bacillus subtilis spore coat protein through formation of the main metabolites AFQ1 and epi-AFQ1.
Subagia R, Schweiger W, Kunz-Vekiru E, Wolfsberger D, Schatzmayr G, Ribitsch D, Guebitz GM. Subagia R, et al. Front Microbiol. 2024 Oct 4;15:1406707. doi: 10.3389/fmicb.2024.1406707. eCollection 2024. Front Microbiol. 2024. PMID: 39430102 Free PMC article. - Impact of food processing and detoxification treatments on mycotoxin contamination.
Karlovsky P, Suman M, Berthiller F, De Meester J, Eisenbrand G, Perrin I, Oswald IP, Speijers G, Chiodini A, Recker T, Dussort P. Karlovsky P, et al. Mycotoxin Res. 2016 Nov;32(4):179-205. doi: 10.1007/s12550-016-0257-7. Epub 2016 Aug 23. Mycotoxin Res. 2016. PMID: 27554261 Free PMC article. Review.
References
- Alberts JF, Engelbrecht Y, Steyn PS, Holzapfel WH, van Zyl WH. Biological degradation of aflatoxin B1 by Rhodococcus erythropolis cultures. Int J Food Microbiol. 2006;109:121–126. - PubMed
- Bashiri G, Squire CJ, Moreland NJ, Baker EN. Crystal structures of F420-dependent glucose-6-phosphate dehydrogenase FGD1 involved in the activation of the anti-tuberculosis drug candidate PA-824 reveal the basis of coenzyme and substrate binding. J Biol Chem. 2008;283:17531–17541. - PubMed
- Biswal BK, Cherney MM, Wang M, Garen C, James MN. Structures of Mycobacterium tuberculosis pyridoxine 5′-phosphate oxidase and its complexes with flavin mononucleotide and pyridoxal 5′-phosphate. Acta Crystallogr D Biol Crystallogr. 2005;61:1492–1499. - PubMed
- Campbell PM, Cao AT, Hines ER, East PD, Gordon KH. Proteomic analysis of the peritrophic matrix from the gut of the caterpillar, Helicoverpa armigera. Insect Biochem Mol Biol. 2008;38:950–958. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases