A complete developmental sequence of a Drosophila neuronal lineage as revealed by twin-spot MARCM - PubMed (original) (raw)
A complete developmental sequence of a Drosophila neuronal lineage as revealed by twin-spot MARCM
Hung-Hsiang Yu et al. PLoS Biol. 2010.
Abstract
Drosophila brains contain numerous neurons that form complex circuits. These neurons are derived in stereotyped patterns from a fixed number of progenitors, called neuroblasts, and identifying individual neurons made by a neuroblast facilitates the reconstruction of neural circuits. An improved MARCM (mosaic analysis with a repressible cell marker) technique, called twin-spot MARCM, allows one to label the sister clones derived from a common progenitor simultaneously in different colors. It enables identification of every single neuron in an extended neuronal lineage based on the order of neuron birth. Here we report the first example, to our knowledge, of complete lineage analysis among neurons derived from a common neuroblast that relay olfactory information from the antennal lobe (AL) to higher brain centers. By identifying the sequentially derived neurons, we found that the neuroblast serially makes 40 types of AL projection neurons (PNs). During embryogenesis, one PN with multi-glomerular innervation and 18 uniglomerular PNs targeting 17 glomeruli of the adult AL are born. Many more PNs of 22 additional types, including four types of polyglomerular PNs, derive after the neuroblast resumes dividing in early larvae. Although different offspring are generated in a rather arbitrary sequence, the birth order strictly dictates the fate of each post-mitotic neuron, including the fate of programmed cell death. Notably, the embryonic progenitor has an altered temporal identity following each self-renewing asymmetric cell division. After larval hatching, the same progenitor produces multiple neurons for each cell type, but the number of neurons for each type is tightly regulated. These observations substantiate the origin-dependent specification of neuron types. Sequencing neuronal lineages will not only unravel how a complex brain develops but also permit systematic identification of neuron types for detailed structure and function analysis of the brain.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
Figure 1. Examples of paired sister clones, the AL glomerular architecture, and a summary of adPNs.
(A–C) GMC clones (magenta) pairs with NB clones (green) of different sizes depending on when mitotic recombination occurred in a protracted lineage. Judging from the size of the accompanying NB clone, one can determine when the GMC of a particular neuron was born in the lineage. One can therefore deduce in the adPN lineage: the VM3-targeting neuron (magenta in [A]) born around the beginning, the 1-targeting neuron (magenta in [B]) derived in the middle, and the DL2v-targeting neuron (magenta in [C]) made near the end. (D) A schematic illustration of an uniglomerular adPN (black) that connects one of the 50 or so AL glomeruli with the calyx (CA) of the mushroom body (MB) and the lateral horn (LH). The relative position of three populations of PNs, the adPNs (blue), lPNs (red), and vPNs (orange), is also shown. (E–H) All identifiable glomeruli in the AL are shown in four anterior-to-posterior focal sections. The glomerular targets of previously identified uniglomerular adPNs, lPNs, and vPNs are labeled in blue, red, and orange, respectively. The glomerular targets of the uniglomerular adPNs identified in this study are shown in cyan. And the glomeruli with yellow labels have not yet found their corresponding uniglomerular PNs. (I) adPNs identified previously and in this study are summarized. The adPNs with known birth order are further arranged with respect to the lineage development. Fly brains were counterstained with nc82 mAb (blue) in this and all other figures, which permits determination of glomerular identity in the AL. The scale bar in this and all other figures equals 10 µm.
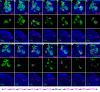
Figure 2. Twelve types of early-larval-derived GH146-positive uniglomerular adPNs.
Twin-spot MARCM clones of adPNs labeled with GAL4-GH146 (A-M) or acj6-GAL4 (N). Top panels: composite confocal images of sister clones in the AL; middle panels: single focal sections of the AL covering the glomerular targets of GMC progeny (magenta); bottom panels: axon projections of GMC progeny (magenta); islets in bottom panels: axon projections of both GMC progeny (magenta) and its paired NB clone (green). Note each adPN type (magenta) consistently pairs with adPN NB clones (green) of specific compositions. Analysis of NB clones revealed the 12 types of GH146-positive adPNs are made in an invariant sequence from (A) to (M). And all the lone, unpaired NB clones (H), whose preceding GMC progeny probably die prematurely, were induced in the interval between VA1d and 1 adPNs. The sequence of early-larval adPN neurogenesis is summarized in the bottom. In addition, there are multiple neurons per type, as evidenced in middle panels that the glomerular target of GMC progeny can be co-labeled by its accompanying NB clone. For the lineage after VA1lm PNs, one can visualize GH146-negative adPNs with acj6-GAL4 as revealed in (N) where the last VA1lm adPN pairs with a 32-neuron-containing NB clone.
Figure 3. Ten types of late-larval-derived Acj6-positive adPNs.
Late-larval-derived twin-spot clones labeled with acj6-GAL4. Top and middle panels: composite confocal images of the AL showing single adPNs only or both single adPNs (magenta) and their paired NB clones (green); bottom panels: the axon projections of single adPNs and in islets the projections of single adPNs (magenta) and their accompanying NB clones (green). Analysis of NB clones paired with distinct adPNs revealed 10 additional adPN types are made in an invariant sequence as summarized in the bottom. Note presence of four types of polyglomerular PNs that exhibit different patterns of AL elaboration while acquiring analogous axon projections in the LH (E–H).
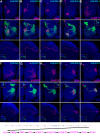
Figure 4. Eighteen types of embryonic-born adPNs.
Embryonic-derived twin-spot clones labeled with GAL4-GH146 (A–V) or acj6-GAL4 (X–Z). Top two panels: composite confocal images of the AL showing single adPNs (magenta) and the paired NB clones (green); middle panels: single focal sections of the AL covering the glomerular targets of single adPNs; bottom two panels: axon projections of single adPNs (magenta) and the accompanying NB clones (green). 22 types of GH146-labeled NB clones can be distinguished and are shown in the order of decreasing complexity from (A) to (V). 15 of them pair with distinct adPNs while seven of them exist alone (no magenta labeling). The unpaired NB clones in (H), (J), and (M), when labeled with acj6-GAL4, were paired with novel GH146-negative adPNs, including one additional type of polyglomerular PN (X–Z). The sequence of embryonic-derived adPNs is shown in (W). Note presence of one neuron per type (except VM3-targeting PNs in [R] and [T]) in the lineage of primary neurons, as evidenced by the glomerular targets of single adPNs (magenta) not innervated by their accompanying NB clones (green).
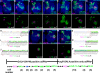
Figure 5. Distinct cell counts in different adPN types.
Twin-spot clones labeled with GAL4-GH146 (A–G) or acj6-GAL4 (K–M). Upper panels: composite confocal images of sister clones in the AL; lower panels: single focal sections showing no innervation of the magenta glomeruli by the green NB clones, indicating clones derived during birth of the last sibling of the preceding adPN type. The clones shown in (A) and illustrated in (H) reveal the adPN NB makes five VA1lm-targeting PNs following derivation of the last DM6-targeting PN. Illustrations of lineage development for additional twin-spot clones are shown in (I), (J), and (N) to (P). Invariant cell counts were obtained for the majority of NB clones paired with the last sibling of the preceding adPN type (see Tables S3 and S4). These support production of a fixed number of neurons for each multi-cellular adPN type, as summarized in the bottom.
Similar articles
- Lineage analysis of Drosophila lateral antennal lobe neurons reveals notch-dependent binary temporal fate decisions.
Lin S, Kao CF, Yu HH, Huang Y, Lee T. Lin S, et al. PLoS Biol. 2012;10(11):e1001425. doi: 10.1371/journal.pbio.1001425. Epub 2012 Nov 20. PLoS Biol. 2012. PMID: 23185131 Free PMC article. - Clonal analysis of Drosophila antennal lobe neurons: diverse neuronal architectures in the lateral neuroblast lineage.
Lai SL, Awasaki T, Ito K, Lee T. Lai SL, et al. Development. 2008 Sep;135(17):2883-93. doi: 10.1242/dev.024380. Epub 2008 Jul 24. Development. 2008. PMID: 18653555 - Drosophila olfactory local interneurons and projection neurons derive from a common neuroblast lineage specified by the empty spiracles gene.
Das A, Sen S, Lichtneckert R, Okada R, Ito K, Rodrigues V, Reichert H. Das A, et al. Neural Dev. 2008 Dec 3;3:33. doi: 10.1186/1749-8104-3-33. Neural Dev. 2008. PMID: 19055770 Free PMC article. - Birth time/order-dependent neuron type specification.
Kao CF, Lee T. Kao CF, et al. Curr Opin Neurobiol. 2010 Feb;20(1):14-21. doi: 10.1016/j.conb.2009.10.017. Epub 2009 Nov 26. Curr Opin Neurobiol. 2010. PMID: 19944594 Free PMC article. Review. - [Progress on cell lineage analysis in Drosophila melanogaster].
Zhang SP, Xue L. Zhang SP, et al. Yi Chuan. 2012 Jul;34(7):819-28. doi: 10.3724/sp.j.1005.2012.00819. Yi Chuan. 2012. PMID: 22805207 Review. Chinese.
Cited by
- Whole-brain annotation and multi-connectome cell typing of Drosophila.
Schlegel P, Yin Y, Bates AS, Dorkenwald S, Eichler K, Brooks P, Han DS, Gkantia M, Dos Santos M, Munnelly EJ, Badalamente G, Serratosa Capdevila L, Sane VA, Fragniere AMC, Kiassat L, Pleijzier MW, Stürner T, Tamimi IFM, Dunne CR, Salgarella I, Javier A, Fang S, Perlman E, Kazimiers T, Jagannathan SR, Matsliah A, Sterling AR, Yu SC, McKellar CE; FlyWire Consortium; Costa M, Seung HS, Murthy M, Hartenstein V, Bock DD, Jefferis GSXE. Schlegel P, et al. Nature. 2024 Oct;634(8032):139-152. doi: 10.1038/s41586-024-07686-5. Epub 2024 Oct 2. Nature. 2024. PMID: 39358521 Free PMC article. - Spatial constraints and cell surface molecule depletion structure a randomly connected learning circuit.
Thornton-Kolbe EM, Ahmed M, Gordon FR, Sieriebriennikov B, Williams DL, Kurmangaliyev YZ, Clowney EJ. Thornton-Kolbe EM, et al. bioRxiv [Preprint]. 2024 Jul 21:2024.07.17.603956. doi: 10.1101/2024.07.17.603956. bioRxiv. 2024. PMID: 39071296 Free PMC article. Preprint. - Multiple isoforms of the Activin-like receptor baboon differentially regulate proliferation and conversion behaviors of neuroblasts and neuroepithelial cells in the Drosophila larval brain.
Lee GG, Peterson AJ, Kim MJ, O'Connor MB, Park JH. Lee GG, et al. PLoS One. 2024 Jun 24;19(6):e0305696. doi: 10.1371/journal.pone.0305696. eCollection 2024. PLoS One. 2024. PMID: 38913612 Free PMC article. - Imp is expressed in INPs and newborn neurons where it regulates neuropil targeting in the central complex.
Munroe JA, Doe CQ. Munroe JA, et al. Neural Dev. 2023 Nov 29;18(1):9. doi: 10.1186/s13064-023-00177-9. Neural Dev. 2023. PMID: 38031099 Free PMC article. - Input density tunes Kenyon cell sensory responses in the Drosophila mushroom body.
Ahmed M, Rajagopalan AE, Pan Y, Li Y, Williams DL, Pedersen EA, Thakral M, Previero A, Close KC, Christoforou CP, Cai D, Turner GC, Clowney EJ. Ahmed M, et al. Curr Biol. 2023 Jul 10;33(13):2742-2760.e12. doi: 10.1016/j.cub.2023.05.064. Epub 2023 Jun 21. Curr Biol. 2023. PMID: 37348501 Free PMC article.
References
- Young R. W. Cell differentiation in the retina of the mouse. Anat Rec. 1985;212:199–205. - PubMed
- Noctor S. C, Flint A. C, Weissman T. A, Dammerman R. S, Kriegstein A. R. Neurons derived from radial glial cells establish radial units in neocortex. Nature. 2001;409:714–720. - PubMed
- Urbach R, Schnabel R, Technau G. M. The pattern of neuroblast formation, mitotic domains and proneural gene expression during early brain development in Drosophila. Development. 2003;130:3589–3606. - PubMed
- Truman J. W, Bate M. Spatial and temporal patterns of neurogenesis in the central nervous system of Drosophila melanogaster. Dev Biol. 1988;125:145–157. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases