HIV-1 Populations in Semen Arise through Multiple Mechanisms - PubMed (original) (raw)
. 2010 Aug 19;6(8):e1001053.
doi: 10.1371/journal.ppat.1001053.
Li-Hua Ping, Oliver Dibben, Cassandra B Jabara, Leslie Arney, Laura Kincer, Yuyang Tang, Marcia Hobbs, Irving Hoffman, Peter Kazembe, Corbin D Jones, Persephone Borrow, Susan Fiscus, Myron S Cohen, Ronald Swanstrom; Center for HIV/AIDS Vaccine Immunology
Affiliations
- PMID: 20808902
- PMCID: PMC2924360
- DOI: 10.1371/journal.ppat.1001053
HIV-1 Populations in Semen Arise through Multiple Mechanisms
Jeffrey A Anderson et al. PLoS Pathog. 2010.
Abstract
HIV-1 is present in anatomical compartments and bodily fluids. Most transmissions occur through sexual acts, making virus in semen the proximal source in male donors. We find three distinct relationships in comparing viral RNA populations between blood and semen in men with chronic HIV-1 infection, and we propose that the viral populations in semen arise by multiple mechanisms including: direct import of virus, oligoclonal amplification within the seminal tract, or compartmentalization. In addition, we find significant enrichment of six out of nineteen cytokines and chemokines in semen of both HIV-infected and uninfected men, and another seven further enriched in infected individuals. The enrichment of cytokines involved in innate immunity in the seminal tract, complemented with chemokines in infected men, creates an environment conducive to T cell activation and viral replication. These studies define different relationships between virus in blood and semen that can significantly alter the composition of the viral population at the source that is most proximal to the transmitted virus.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
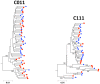
Figure 1. Neighbor-joining tree of SGA-derived env amplicons for two patients with HIV-1 subtype C demonstrating equilibration between blood and semen.
Blood SGA-env sequences (red circles) and semen SGA-env sequences (blue triangles). Bootstrap values ≥70 are shown. Genetic distance is indicated at the bottom of the figure, and represents the number of nucleotide substitutions per site. An outgroup was included to root the tree but is not shown.
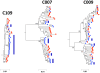
Figure 2. Neighbor-joining tree of SGA-derived env amplicons for three patients with HIV-1 subtype C demonstrating clonal amplification of identical or nearly identical sequences within semen.
Blood SGA-env sequences (red circles) and semen SGA-env sequences (blue triangles). Vertical blue bars highlight clonal amplification of specific variants. Bootstrap values ≥70 are shown. An outgroup was included to root the tree but is not shown.
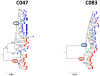
Figure 3. Neighbor-joining tree of SGA-derived env amplicons for two patients with HIV-1 subtype C demonstrating compartmentalization between blood and semen.
Blood SGA-env sequences (red circles) and semen SGA-env sequences (blue triangles). Ovals highlight clades of compartmentalized sequences in the semen or blood. Vertical blue bars highlight clonal amplification of specific variants. Bootstrap values ≥70 are shown. An outgroup was included to root the tree but is not shown.
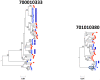
Figure 4. Neighbor-joining tree of SGA-derived env amplicons for two patients with HIV-1 subtype B demonstrating amplification and equilibration in the left panel, and amplification and compartmentalization of seminal sequences in the right panel.
Blood SGA-env sequences (red circles) and semen SGA-env sequences (blue triangles). The oval highlights clades of compartmentalized sequences in the semen. Vertical blue bars highlight clonal amplification of specific variants. Bootstrap values ≥70 are shown. An outgroup was included to root the tree but is not shown.
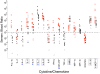
Figure 5. Scatter plot of semen:blood analyte ratios of 17 cytokines and chemokines in HIV-1 infected (red circles) (n = 12), uninfected Malawi men (black circles) (n = 6), and uninfected US men (black triangles) (n = 12).
Values that were below the lower limit of detection were reported as the mid-point between the lower level of detection and zero. Semen:blood ratios were excluded if both compartments were below the level of detection; thus, IL12 and IL13 were not included in the analysis. In the infected men, there were 6 to 11 subjects with semen:blood analyte ratios for IL-1b, IL-2, IL-4, and IFN-g, whereas the remaining cytokines and chemokines had data from all 12 infected men. In addition, after exclusion of values below the limit of detection in both semen and blood compartments, we quantified the semen:blood ratio for each cytokine and chemokine from a range of 10 to 17 HIV-1 uninfected men. The dashed line illustrates an equivalent semen:blood analyte ratio. Horizontal lines represent median values. Blue asterisks denote significant differences between infected and uninfected men, P value from Mann-Whitney test <0.003 (to correct for multiple comparisons). Boxed cytokines/chemokines have median semen:blood analyte ratios >5 in both HIV-1 infected and uninfected men.
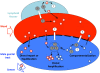
Figure 6. Model demonstrating HIV-1 populations in the blood and male genital tract.
Target cells traffic from blood to the male genital tract. A semi-permeable barrier separates blood from the genital tract that allows passage of some free virus and uninfected and infected cells. Equilibration of blood and seminal plasma sequences occurs through direct import of sequences from blood to semen. Clonal amplification results from establishment of a local focus of infection of anatomically isolated uninfected cells in the genital tract. Compartmentalization occurs when resident cell populations in the male genital tract become infected and there is persistent local replication. Gray circle, HIV-1 infected target cell. White circle, HIV-1 uninfected target cell. Red circle, blood-derived virus. Blue circle, male genital tract-derived virus.
Similar articles
- Deep Sequencing Reveals Compartmentalized HIV-1 in the Semen of Men with and without Sexually Transmitted Infection-Associated Urethritis.
Council OD, Zhou S, McCann CD, Hoffman I, Tegha G, Kamwendo D, Matoga M, Kosakovsky Pond SL, Cohen MS, Swanstrom R. Council OD, et al. J Virol. 2020 Jun 1;94(12):e00151-20. doi: 10.1128/JVI.00151-20. Print 2020 Jun 1. J Virol. 2020. PMID: 32269129 Free PMC article. - Higher viral load and genetic diversity of HIV-1 in seminal compartments than in blood of seven Chinese men who have sex with men and have early HIV-1 infection.
Jiao YM, Chen GL, Zhu WJ, Huang HH, Fu JL, Chen WW, Shi M, Zhang T, Wu H, Wang FS. Jiao YM, et al. Microbiol Immunol. 2017 Jun;61(6):239-246. doi: 10.1111/1348-0421.12488. Microbiol Immunol. 2017. PMID: 28500746 - Compartmentalization and Clonal Amplification of HIV-1 in the Male Genital Tract Characterized Using Next-Generation Sequencing.
Kariuki SM, Selhorst P, Anthony C, Matten D, Abrahams MR, Martin DP, Ariën KK, Rebe K, Williamson C, Dorfman JR. Kariuki SM, et al. J Virol. 2020 Jun 1;94(12):e00229-20. doi: 10.1128/JVI.00229-20. Print 2020 Jun 1. J Virol. 2020. PMID: 32269124 Free PMC article. - Assays for the determination of HIV-1 load in semen: a review of indications, methods and performance in vitro.
Chan DJ, McNally L. Chan DJ, et al. Curr HIV Res. 2008 May;6(3):182-8. doi: 10.2174/157016208784324949. Curr HIV Res. 2008. PMID: 18473781 Review. - Seminal exosomes and HIV-1 transmission.
Ouattara LA, Anderson SM, Doncel GF. Ouattara LA, et al. Andrologia. 2018 Dec;50(11):e13220. doi: 10.1111/and.13220. Andrologia. 2018. PMID: 30569645 Free PMC article. Review.
Cited by
- Update on known and emergent viruses affecting human male genital tract and fertility.
Dabizzi S, Maggi M, Torcia MG. Dabizzi S, et al. Basic Clin Androl. 2024 Mar 14;34(1):6. doi: 10.1186/s12610-024-00222-5. Basic Clin Androl. 2024. PMID: 38486154 Free PMC article. Review. - Are sexually transmitted infections associated with male infertility? A systematic review and in-depth evaluation of the evidence and mechanisms of action of 11 pathogens.
Khalafalla K, El Ansari W, Sengupta P, Majzoub A, Elbardisi H, Canguven O, El-Ansari K, Arafa M. Khalafalla K, et al. Arab J Urol. 2023 Jul 5;21(4):216-232. doi: 10.1080/2090598X.2023.2218566. eCollection 2023. Arab J Urol. 2023. PMID: 38178949 Free PMC article. Review. - EFdA efficiently suppresses HIV replication in the male genital tract and prevents penile HIV acquisition.
Kovarova M, Wessel SE, Johnson CE, Anderson SV, Cottrell ML, Sykes C, Cohen MS, Garcia JV. Kovarova M, et al. mBio. 2023 Aug 31;14(4):e0222422. doi: 10.1128/mbio.02224-22. Epub 2023 Jun 12. mBio. 2023. PMID: 37306625 Free PMC article. - Viral Infections and Male Infertility: A Comprehensive Review of the Role of Oxidative Stress.
Akhigbe RE, Dutta S, Hamed MA, Ajayi AF, Sengupta P, Ahmad G. Akhigbe RE, et al. Front Reprod Health. 2022 Feb 3;4:782915. doi: 10.3389/frph.2022.782915. eCollection 2022. Front Reprod Health. 2022. PMID: 36303638 Free PMC article. Review. - Human seminal virome: a panel based on recent literature.
de Albuquerque BHDR, de Oliveira MTFC, Aderaldo JF, de Medeiros Garcia Torres M, Lanza DCF. de Albuquerque BHDR, et al. Basic Clin Androl. 2022 Sep 6;32(1):16. doi: 10.1186/s12610-022-00165-9. Basic Clin Androl. 2022. PMID: 36064315 Free PMC article. Review.
References
- Pilcher CD, Price MA, Hoffman IF, Galvin S, Martinson FE, et al. Frequent detection of acute primary HIV infection in men in Malawi. AIDS. 2004;18:517–524. - PubMed
- Pilcher CD, Shugars DC, Fiscus SA, Miller WC, Menezes P, et al. HIV in body fluids during primary HIV infection: implications for pathogenesis, treatment and public health. AIDS. 2001;15:837–845. - PubMed
- Tindall B, Evans L, Cunningham P, McQueen P, Hurren L, et al. Identification of HIV-1 in semen following primary HIV-1 infection. AIDS. 1992;6:949–952. - PubMed
- Pullium JK, Adams DR, Jackson E, Kim CN, Smith DK, et al. Pig-tailed macaques infected with human immunodeficiency virus (HIV) type 2GB122 or simian/HIV89.6p express virus in semen during primary infection: new model for genital tract shedding and transmission. J Infect Dis. 2001;183:1023–1030. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P30 AI050410/AI/NIAID NIH HHS/United States
- R37 DK049381/DK/NIDDK NIH HHS/United States
- U01 AI067854/AI/NIAID NIH HHS/United States
- P30 AI50410/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases