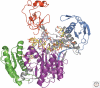Ancestral roles of small RNAs: an Ago-centric perspective - PubMed (original) (raw)
Review
Ancestral roles of small RNAs: an Ago-centric perspective
Leemor Joshua-Tor et al. Cold Spring Harb Perspect Biol. 2011.
Abstract
RNAi has existed at least since the divergence of prokaryotes and eukaryotes. This collection of pathways responds to a diversity of "abberant" RNAs and generally silences or eliminates genes sharing sequence content with the silencing trigger. In the canonical pathway, double-stranded RNAs are processed into small RNAs, which guide effector complexes to their targets by complementary base pairing. Many alternative routes from silencing trigger to small RNA are continuously being uncovered. Though the triggers of the pathway and the mechanisms of small RNA production are many, all RNAi-related mechanisms share Argonaute proteins as the heart of their effector complexes. These can act as self-contained silencing machines, binding directly to small RNAs, carrying out homology-based target recognition, and in some cases cleaving targets using an endogenous nuclease domain. Here, we discuss the diversity of Argonaute proteins from a structural and functional perspective.
Figures
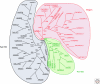
Figure 1.
The three clades of the Argonautes. The Ago-like clade, shown in black, is based on similarity to AtAgo1; the Piwi-like clade, shown in green based on similarity to DmPiwi and the Wago's, which is a worm-specific clade is shown in red. Argonautes that contain a complete catalytic motif are underlined (Adapted from Tolia and Joshua-Tor 2007).
Figure 2.
The crystal structure of Argonaute from P. furiosus shown as a ribbon diagram. The amino-terminal domain is in blue, the PAZ domain is red, the middle domain is green, and the PIWI domain is in purple. The interdomain connector is shown in yellow. A close-up of the active site residues coordinating a Mn++ ion (pink ball) is shown in the inset.
Figure 3.
The crystal structure of Argonaute from Thermus thermophilus in complex with a 21-nucleotide DNA guide (in stick with gray carbons) and a 12-nucleotide RNA target (in stick with yellow carbons). The amino-terminal domain is in blue, the PAZ domain in red, the mid domain in green and the PIWI domain in purple. The two linkers are shown in gray. The first 12 nucleotides and the two 3′-nucleotides of the DNA guide are observed in the structure. The 3′ nucleotides are bound to the PAZ domain.
Figure 4.
The crystal structure of Argonaute from T. thermophilus in complex with a 21-nucleotide DNA guide (in stick with gray carbons) and a 19-nucleotide RNA target (in stick with yellow carbons). Domains are colored as in Figure 3. The first 16 nucleotides of the DNA guide and 15 nucleotides of the RNA target strand are observed in the structure. The 3′-end nucleotides of the guide are no longer bound to the PAZ domain in this structure.
Similar articles
- RISC assembly: Coordination between small RNAs and Argonaute proteins.
Kobayashi H, Tomari Y. Kobayashi H, et al. Biochim Biophys Acta. 2016 Jan;1859(1):71-81. doi: 10.1016/j.bbagrm.2015.08.007. Epub 2015 Aug 22. Biochim Biophys Acta. 2016. PMID: 26303205 Review. - Kinetic Analysis of Target RNA Binding and Slicing by Human Argonaute 2 Protein.
Willkomm S, Restle T. Willkomm S, et al. Methods Mol Biol. 2017;1517:277-290. doi: 10.1007/978-1-4939-6563-2_19. Methods Mol Biol. 2017. PMID: 27924489 - Lessons on RNA silencing mechanisms in plants from eukaryotic argonaute structures.
Poulsen C, Vaucheret H, Brodersen P. Poulsen C, et al. Plant Cell. 2013 Jan;25(1):22-37. doi: 10.1105/tpc.112.105643. Epub 2013 Jan 9. Plant Cell. 2013. PMID: 23303917 Free PMC article. - Single-molecule fluorescence measurements reveal the reaction mechanisms of the core RISC, composed of human Argonaute 2 and a guide RNA.
Jo MH, Song JJ, Hohng S. Jo MH, et al. BMB Rep. 2015 Dec;48(12):643-4. doi: 10.5483/bmbrep.2015.48.12.235. BMB Rep. 2015. PMID: 26592935 Free PMC article. - RNA-induced initiation of transcriptional silencing (RITS) complex structure and function.
Bhattacharjee S, Roche B, Martienssen RA. Bhattacharjee S, et al. RNA Biol. 2019 Sep;16(9):1133-1146. doi: 10.1080/15476286.2019.1621624. Epub 2019 Jun 18. RNA Biol. 2019. PMID: 31213126 Free PMC article. Review.
Cited by
- Prokaryotic Argonaute nuclease cooperates with co-encoded RNase to acquire guide RNAs and target invader DNA.
Agapov A, Panteleev V, Kropocheva E, Kanevskaya A, Esyunina D, Kulbachinskiy A. Agapov A, et al. Nucleic Acids Res. 2024 Jun 10;52(10):5895-5911. doi: 10.1093/nar/gkae345. Nucleic Acids Res. 2024. PMID: 38716875 Free PMC article. - Leishmania infection upregulates and engages host macrophage Argonaute 1, and system-wide proteomics reveals Argonaute 1-dependent host response.
Moradimotlagh A, Chen S, Koohbor S, Moon KM, Foster LJ, Reiner N, Nandan D. Moradimotlagh A, et al. Front Immunol. 2023 Nov 30;14:1287539. doi: 10.3389/fimmu.2023.1287539. eCollection 2023. Front Immunol. 2023. PMID: 38098491 Free PMC article. - Themes and variations on piRNA-guided transposon control.
Loubalova Z, Konstantinidou P, Haase AD. Loubalova Z, et al. Mob DNA. 2023 Sep 2;14(1):10. doi: 10.1186/s13100-023-00298-2. Mob DNA. 2023. PMID: 37660099 Free PMC article. Review. - Enzymatic reactions of AGO4 in RNA-directed DNA methylation: siRNA duplex loading, passenger strand elimination, target RNA slicing, and sliced target retention.
Wang F, Huang HY, Huang J, Singh J, Pikaard CS. Wang F, et al. Genes Dev. 2023 Feb 1;37(3-4):103-118. doi: 10.1101/gad.350240.122. Epub 2023 Feb 6. Genes Dev. 2023. PMID: 36746605 Free PMC article. - An introduction to PIWI-interacting RNAs (piRNAs) in the context of metazoan small RNA silencing pathways.
Haase AD. Haase AD. RNA Biol. 2022 Jan;19(1):1094-1102. doi: 10.1080/15476286.2022.2132359. RNA Biol. 2022. PMID: 36217279 Free PMC article. Review.
References
- Alexander WG, Raju NB, Xiao H, Hammond TM, Perdue TD, Metzenberg RL, Pukkila PJ, Shiu PK 2008. DCL-1 colocalizes with other components of the MSUD machinery and is required for silencing. Fungal Genet Biol 45: 719–727 - PubMed
- Aravin AA, Hannon GJ, Brennecke J 2007. The Piwi-piRNA pathway provides an adaptive defense in the transposon arms race. Science 318: 761–764 - PubMed
- Bernstein E, Caudy AA, Hammond SM, Hannon GJ 2001. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409: 363–366 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources