Ubiquitin-mediated mRNP dynamics and surveillance prior to budding yeast mRNA export - PubMed (original) (raw)
Ubiquitin-mediated mRNP dynamics and surveillance prior to budding yeast mRNA export
Nahid Iglesias et al. Genes Dev. 2010.
Abstract
The evolutionarily conserved mRNA export receptor Mex67/NXF1 associates with mRNAs through its adaptor, Yra1/REF, allowing mRNA ribonucleoprotein (mRNP) exit through nuclear pores. However, alternate adaptors should exist, since Yra1 is dispensable for mRNA export in Drosophila and Caenorhabditis elegans. Here we report that Mex67 interacts directly with Nab2, an essential shuttling mRNA-binding protein required for export. We further show that Yra1 enhances the interaction between Nab2 and Mex67, and becomes dispensable in cells overexpressing Nab2 or Mex67. These observations appoint Nab2 as a potential adaptor for Mex67, and define Yra1/REF as a cofactor stabilizing the adaptor-receptor interaction. Importantly, Yra1 ubiquitination by the E3 ligase Tom1 promotes its dissociation from mRNP before export. Finally, loss of perinuclear Mlp proteins suppresses the growth defects of Tom1 and Yra1 ubiquitination mutants, suggesting that Tom1-mediated dissociation of Yra1 from Nab2-bound mRNAs is part of a surveillance mechanism at the pore, ensuring export of mature mRNPs only.
Figures
Figure 1.
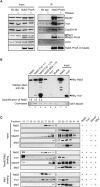
Nab2, Yra1, and Mex67 form a trimeric complex. (A) Nab2 coimmunoprecipitates Mex67 and Yra1 in vivo in an RNA-independent manner. Yeast extracts from Nab2-ProtA or nontagged (No tag) cells and expressing HA-tagged Cbp20 were treated (+) or not (−) with RNases and purified with IgG-Sepharose beads. Input and eluted fractions (IP) were analyzed by Western blotting with anti-Mex67, anti-Yra1, anti-HA, anti-Rap1, anti-Actin, and anti-ProtA antibodies. (B) Yra1 promotes binding of Nab2 and Mex67. GST-Mex67 on beads was incubated with purified His6-Nab2 (lane 1), His6-Nab2 plus equimolar (lane 2) or 10× molar (lane 3) amounts of purified His6-Yra1, BSA (lane 4), purified His6-Yra1 alone (lane 5), or buffer alone (lane 6). His6-Nab2 and His6-Yra1 input are in lanes 7 and 8, respectively. Western blot (top panel) with anti-His antibodies and Coomassie staining for GST-Mex67 (bottom panel) are shown. Quantification of the signal observed for His6-Nab2 association with GST-Mex67 is indicated below. (C) Size fractionation of purified Mex67/Mtr2, Nab2, and Yra1 alone or in complex. His6-Nab2-, His6-Yra1-, His6-Mtr2/Mex67-, or His6-Mtr2-purified proteins were loaded on S200 sizing columns alone or after complex assembly of His6-Mtr2/Mex67 or His6-Mtr2 with His6-Nab2 and His6-Yra1 as indicated on the right. (Top numbers) Collected fractions were analyzed by Western blotting with antibodies specific for Nab2, Yra1, Mex67, and Histidine to detect Mtr2.
Figure 2.
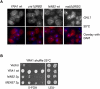
Nab2 and Yra1 are part of the same mRNA export pathway. (A) FISH analysis of GAL1 mRNA in the indicated cells induced with galactose for 30 min at 30°C. DAPI staining or FISH/DAPI overlays are shown below each panel to localize the RNA signal with respect to the nucleus. (B) Δ_yra1_ is bypassed by overexpressing NAB2 or MEX67. The Yra1 shuffle strain transformed with the indicated plasmids was spotted as 10-fold serial dilutions onto 5-FOA to select against the wild-type pURA3-YRA1 plasmid, or onto a selective plate to control for cell number (last two dilutions only). (2μ) High-copy plasmids.
Figure 3.
Yra1 is ubiquitinated by Tom1. (A) HA-YRA1 cDNA-shuffled (lanes 2,3) and Δ_tom1_ HA-YRA1 cDNA-shuffled (lane 4) strains containing a PCUP1_∷_HIS6-ubiquitin plasmid (+) or an empty vector (−). His6-Ub-conjugated forms purified on nickel column (Ni+ column) were examined with their corresponding total input extracts by Western blotting with anti-His, anti-HA (Yra1), anti-Nab2, and anti-Mex67 antibodies. (Lane 2) The empty vector allowed us to control for background binding to the nickel column. (B) Tom1 does not affect Yra1 turnover. Wild-type (wt) and Δ_tom1_ cells were transformed with a plasmid expressing HA-Yra1 from a galactose (GAL)-inducible promoter. Cells were exponentially grown in galactose. At time 0, cultures were shifted to 37°C to exacerbate the potential instability, and HA-Yra1 expression was shut off by the addition of 2% glucose. Yra1 turnover was followed by collecting protein samples at the indicated time points in glucose and Western blotting with antibodies against HA or the C-terminal domain of RNA polymerase II (RNA Pol II) to confirm equal loading. (C) Yra1 domain organization and binding partners, and position of the 22 lysines (black dots). (D) Yra1 ubiquitination in Yra1 wild-type or KR mutant strains. His6-Ub-conjugated forms of HA-Yra1 from YRA1-shuffled (lanes 2,3,5,7) or Δ_tom1_ YRA1-shuffled (lanes 4,6,8) strains expressing the indicated wild-type and Yra1 KR mutant plasmids were purified and analyzed as in A.
Figure 4.
Yra1 ubiquitination reduces interaction with mRNP components. (A) The Yra1 KR9–22 lysine mutant increases the association of Yra1 with nuclear mRNPs. Total extracts (Input) or IgG-Sepharose-purified extracts (IP) from Cbp20-TAP-tagged or nontagged (No tag) strains expressing wild-type or KR9–22 mutant Yra1 were analyzed by Western blotting with anti-Nab2, anti-Mex67, and anti-HA antibodies to detect Yra1. Bead-associated Cbp20-TAP was revealed with an anti-ProtA antibody, and served as control for equal amounts of mRNP pulled down. (B) Tom1-mediated ubiquitination of Yra1 induces Yra1 release from Nab2–Mex67 complexes. Total extracts prepared from Nab2 ProtA-tagged or nontagged (no tag) strains transformed with UbK48R,G76A or empty vector (−) were purified on IgG beads. Western blotting of total extract (Input) or high-salt eluted proteins (IP) were examined with antibodies against Mex67, Yra1, and ProtA. Quantification of the signal observed in the Nab2-ProtA immunoprecipitate for Mex67 and Yra1 (normalized to the signal in wild-type cells transformed with empty vector) is indicated below.
Figure 5.
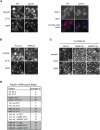
Yra1 ubiquitination is required for efficient mRNA export. (A) Loss of Tom1 interferes with mRNA export. (Left panel) Indicated strains were grown at 25°C, shifted for 3 h to 37°C, and analyzed for poly(A)+ RNA distribution with an oligo (dT)50 probe. (Right panel) Cells were induced with galactose for 30 min at 30°C, and GAL1 mRNA distribution was analyzed by FISH using specific probes. DAPI staining or FISH/DAPI overlays are shown below each panel to localize the RNA signal with respect to the nucleus. (B) Loss of Yra1 ubiquitination interferes with mRNA export. Yra1 wild type and Yra1 KR9–22 cells were grown at 25°C, shifted for 1 h to 37°C, and analyzed for poly(A)+ RNA distribution with an oligo (dT)50 probe. (C) Overexpression of Nab2 and Mex67 suppresses the Yra1 ubiquitination defect. Yra1 KR9–22 cells transformed with high-copy NAB2, MEX67 plasmids, or empty vector were grown at 25°C and analyzed for poly(A)+ RNA distribution as in B. (D) Quantification of FISH analyses: The average percent of cells with an mRNA export defect was determined by examining at least 100 DAPI-stained cells from three independent FISH experiments (except for wild type and Δ_tom1_, the values of which correspond to one experiment). Note that wild type is W303, the corresponding wild type for Δ_tom1_, while Yra1 wild type is YRA1-shuffled + YRA1 wild-type cDNA, the corresponding wild-type for KR9–22.
Figure 6.
Yra1 ubiquitination may be part of an mRNA export surveillance mechanism associated with NPCs. (A) Loss of Mlp2 suppresses the Yra1 KR mutant temperature-sensitive phenotypes. Tenfold serial dilutions of the indicated YRA1-shuffled strains were spotted onto YEPD plates and grown at the indicated temperatures. (B) Loss of Mlp2 suppresses the Δ_tom1_ growth defect. Indicated strains were analyzed as in A. (C) Loss of Rrp6 enhances the growth defect of Δ_tom1_. Indicated strains were analyzed as in A. (D) Model showing the ubiquitin-dependent release of Yra1 from Mlp-retained mRNP complexes, allowing their nuclear exit. Ubiquitination is mediated by Tom1 and another still unidentified E3 ligase (not shown).
Similar articles
- The DEAD-box protein Dbp2 functions with the RNA-binding protein Yra1 to promote mRNP assembly.
Ma WK, Cloutier SC, Tran EJ. Ma WK, et al. J Mol Biol. 2013 Oct 23;425(20):3824-38. doi: 10.1016/j.jmb.2013.05.016. Epub 2013 May 28. J Mol Biol. 2013. PMID: 23721653 Free PMC article. - Perinuclear Mlp proteins downregulate gene expression in response to a defect in mRNA export.
Vinciguerra P, Iglesias N, Camblong J, Zenklusen D, Stutz F. Vinciguerra P, et al. EMBO J. 2005 Feb 23;24(4):813-23. doi: 10.1038/sj.emboj.7600527. Epub 2005 Feb 3. EMBO J. 2005. PMID: 15692572 Free PMC article. - Functional significance of the interaction between the mRNA-binding protein, Nab2, and the nuclear pore-associated protein, Mlp1, in mRNA export.
Fasken MB, Stewart M, Corbett AH. Fasken MB, et al. J Biol Chem. 2008 Oct 3;283(40):27130-43. doi: 10.1074/jbc.M803649200. Epub 2008 Aug 5. J Biol Chem. 2008. PMID: 18682389 Free PMC article. - Ratcheting mRNA out of the nucleus.
Stewart M. Stewart M. Mol Cell. 2007 Feb 9;25(3):327-30. doi: 10.1016/j.molcel.2007.01.016. Mol Cell. 2007. PMID: 17289581 Review. - Structure-function relationships in the Nab2 polyadenosine-RNA binding Zn finger protein family.
Fasken MB, Corbett AH, Stewart M. Fasken MB, et al. Protein Sci. 2019 Mar;28(3):513-523. doi: 10.1002/pro.3565. Epub 2019 Jan 16. Protein Sci. 2019. PMID: 30578643 Free PMC article. Review.
Cited by
- H2B ubiquitylation modulates spliceosome assembly and function in budding yeast.
Hérissant L, Moehle EA, Bertaccini D, Van Dorsselaer A, Schaeffer-Reiss C, Guthrie C, Dargemont C. Hérissant L, et al. Biol Cell. 2014 Apr;106(4):126-38. doi: 10.1111/boc.201400003. Epub 2014 Feb 25. Biol Cell. 2014. PMID: 24476359 Free PMC article. - Nab2 functions in the metabolism of RNA driven by polymerases II and III.
González-Aguilera C, Tous C, Babiano R, de la Cruz J, Luna R, Aguilera A. González-Aguilera C, et al. Mol Biol Cell. 2011 Aug 1;22(15):2729-40. doi: 10.1091/mbc.E11-01-0055. Epub 2011 Jun 16. Mol Biol Cell. 2011. PMID: 21680710 Free PMC article. - Emerging mechanisms of mRNP remodeling regulation.
Chen CY, Shyu AB. Chen CY, et al. Wiley Interdiscip Rev RNA. 2014 Sep-Oct;5(5):713-22. doi: 10.1002/wrna.1241. Epub 2014 Jun 12. Wiley Interdiscip Rev RNA. 2014. PMID: 24923990 Free PMC article. Review. - Automethylation-induced conformational switch in Clr4 (Suv39h) maintains epigenetic stability.
Iglesias N, Currie MA, Jih G, Paulo JA, Siuti N, Kalocsay M, Gygi SP, Moazed D. Iglesias N, et al. Nature. 2018 Aug;560(7719):504-508. doi: 10.1038/s41586-018-0398-2. Epub 2018 Jul 23. Nature. 2018. PMID: 30051891 Free PMC article. - Nuclear export of messenger RNA.
Katahira J. Katahira J. Genes (Basel). 2015 Mar 31;6(2):163-84. doi: 10.3390/genes6020163. Genes (Basel). 2015. PMID: 25836925 Free PMC article. Review.
References
- Bruhn L, Munnerlyn A, Grosschedl R 1997. ALY, a context-dependent coactivator of LEF-1 and AML-1, is required for TCRα enhancer function. Genes Dev 11: 640–653 - PubMed
- Duncan K, Umen JG, Guthrie C 2000. A putative ubiquitin ligase required for efficient mRNA export differentially affects hnRNP transport. Curr Biol 10: 687–696 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases