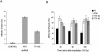A genetic screen identifies the Triple T complex required for DNA damage signaling and ATM and ATR stability - PubMed (original) (raw)
A genetic screen identifies the Triple T complex required for DNA damage signaling and ATM and ATR stability
Kristen E Hurov et al. Genes Dev. 2010.
Abstract
In response to DNA damage, cells activate a complex signal transduction network called the DNA damage response (DDR). To enhance our current understanding of the DDR network, we performed a genome-wide RNAi screen to identify genes required for resistance to ionizing radiation (IR). Along with a number of known DDR genes, we discovered a large set of novel genes whose depletion leads to cellular sensitivity to IR. Here we describe TTI1 (Tel two-interacting protein 1) and TTI2 as highly conserved regulators of the DDR in mammals. TTI1 and TTI2 protect cells from spontaneous DNA damage, and are required for the establishment of the intra-S and G2/M checkpoints. TTI1 and TTI2 exist in multiple complexes, including a 2-MDa complex with TEL2 (telomere maintenance 2), called the Triple T complex, and phosphoinositide-3-kinase-related protein kinases (PIKKs) such as ataxia telangiectasia-mutated (ATM). The components of the TTT complex are mutually dependent on each other, and act as critical regulators of PIKK abundance and checkpoint signaling.
Figures
Figure 1.
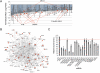
A genome-wide RNAi screen to search for novel DDR genes. (A) Schematic of the pool-based shRNA screen for regulators of the DNA DSB response in mammalian cells. shRNA pools were deconvoluted using microarray hybridization. (B) The screen behavior of a representative shRNA pool of 12,965 shRNAs. shRNAs are graphed based on their mean-normalized log2 Cy5/Cy3 ratio, with a ratio of less than −1 indicating a decrease in the relative abundance of the shRNA greater than twofold, and a ratio of >1 representing an increase in the relative abundance of the shRNA greater than twofold. The population of shRNAs causing IR sensitivity is shaded in red, and IR resistance is shaded in green. (C,D) Gene ontology analysis of the candidate genes required for IR resistance. (C) Significance refers to the −log(_P_-value) determined by the right-tailed Fisher's exact test. Threshold is at 1.25 = −log (P = 0.05). The analysis includes 760 genes from Supplemental Table S1 whose gene symbols were identified by Ingenuity's Knowledge Base. (D) The candidate genes required for IR resistance were classified into several biological processes by the PANTHER program. The analysis includes 760 genes from Supplemental Table S1 whose Entrez Gene IDs were identified by PANTHER.
Figure 2.
Many known DDR, DNA replication, and cell cycle genes scored in the IR sensitivity screen. (A) Screen data of genes that scored with multiple shRNAs, which included known DDR genes, ATM, DNAPK, UBC13, and NBA1. In addition, multiple genes not previously linked to the DDR scored with multiple shRNAs. These data are from the IR screen in U2OS cells treated with 5 Gy and hybridized UNTREATED-END versus IR-END. (B) Ingenuity pathway analysis of genes implicated previously in the DDR, DNA replication, and the cell cycle. Genes labeled in red scored in the IR sensitivity screen (data from multiple screen conditions included) (see the Materials and Methods; Supplemental Table S3). The cross represents a kinase, the squiggle represents an enzyme, the barbell represents a transcription regulator, and the circle represents a protein not classified into one the these three groups. (C) Validation of known DDR, DNA replication, and cell cycle genes using an independent viability assay. MCA was performed using U2OS cells expressing dsRed and the individual shRNAs indicated. Cells were mixed equally with uncolored U2OS cells expressing a control FF shRNA, treated with 3 Gy of IR, and incubated for 7 d, and the relative cell number was determined by FACS. IR-treated samples were normalized to untreated cells and graphed as “percent relative survival.” V2HS numbers (Open Biosystems) for these shRNAs are listed in Supplemental Table S6.
Figure 3.
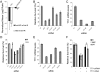
KIAA0406/TTI1 is required for DNA damage resistance. (A) Identification of KIAA0406/TTI1 as a gene required for IR resistance. shRNAs were graphed based on their mean-normalized log2 Cy5/Cy3 ratio. An unrelated shRNA that was considered unchanged is shown for comparison. (B) Validation of the TTI1 shRNA that scored in the screen (#3) and two additional TTI1 shRNAs. Cell viability was monitored using MCA as described in Figure 2C, and was normalized to the FF control virus. Viability in response to 3 Gy of IR was monitored. Error bars represent the standard deviation of three replicates. (C) Quantitative RT–PCR was used to measure the efficiency of shRNA depletion of TTI1 mRNA relative to β-actin as described in the Materials and Methods. Error bars represent the standard deviation of four replicates. (D) siRNAs that target distinct regions of the TTI1 sequence also lead to increased IR sensitivity. Four individual siRNAs from Dharmacon targeting TTI1 were tested (Supplemental Table S7). Cell viability using the MCA assay was monitored as in Figure 2C. Viability in response to 3 Gy of IR or 5 J/m2 UV was monitored. Error bars represent the standard deviation of three replicates. (E) Quantitative RT–PCR was used to measure the efficiency of siRNA depletion of TTI1 mRNA as in C. (F) Expression of a shRNA-resistant TTI1 cDNA restores DNA damage resistance. U2OS cells stably expressing MSCV-HA-Flag-empty vector or MSCV-HA-Flag-TTI1 were infected with retroviruses that express either FF control shRNA or TTI1 shRNA #3, and, after 2 d, were mixed with control cells for MCA. Viability in response to 3 Gy of IR, 75 nM MMC, or 5 nM CPT was monitored. Error bars represent the standard deviation of three replicates.
Figure 4.
Depletion of TTI1 leads to cell cycle checkpoint defects. (A) U2OS cells depleted of TTI1 have a G2/M cell cycle checkpoint defect. U2OS cells stably expressing control, ATR, or TTI1 shRNAs were treated with 10 Gy of IR or left untreated. After 1 h, nocodozole was added, and cells were incubated for an additional 17 h. Cells were fixed and stained for phospho-S10 histone H3. The mean percentage of phospho-S10 histone H3-positive cells (mitotic index) is plotted. The error bars represent standard deviation across three technical replicates. (B) U2OS cells depleted of TTI1 display an intra-S-phase cell cycle checkpoint defect. U2OS cells stably expressing either control, ATM, or TTI1 shRNAs were labeled with 14C-thymidine for 24 h, followed by 24 h in medium containing no label. Cells were treated with 15 Gy of IR for 45, 90, or 180 min; pulse-labeled with 3H-thymidine for 20 min; and harvested; the DNA was fixed onto filters; and 14C and 3H incorporation was measured and normalized to untreated cells. Error bars represent the standard deviation across three technical replicates.
Figure 5.
TTI1 interacts physically with PIKKs to regulate their abundance. (A) TTI1 depletion leads to defective DDR signaling. U2OS cells stably expressing a control, ATM, or two different TTI1 shRNAs were treated with 10 Gy of IR, followed by 4 h of incubation prior to harvesting. Protein lysates were immunoblotted using the indicated antibodies. (B) Protein from U2OS cells stably expressing a control or two different TTI1 shRNAs was immunoblotted using TTI1, ATM, ATR, DNAPK, mTOR, SMG1, TRRAP, or VINCULIN (loading control) antibodies. (C) 293T cells were transiently transfected with Flag-HA-TTI1 or an empty vector for 36 h, and were either left untreated or treated with 10 Gy of IR for 4 h. Proteins immunoprecipitated with HA antibodies were immunoblotted with the indicated antisera. All lanes were run on the same gel and have the same immunoblot exposure. (D) An antibody generated against the C terminus of TTI1 immunoprecipitated endogenous TTI1 and ATM. Normal rabbit IgG was used for a control immunoprecipitation. (E) An antibody against endogenous TTI1 immunoprecipitates endogenous ATR. 293T cells were left untreated or treated with 10 Gy of IR for 4 h. (F) An antibody against endogenous ATM immunoprecipitates endogenous TTI1.
Figure 6.
Human TTI1 and TTI2 associate physically with TEL2. (A) Epitope-tagged TTI1 can coimmunoprecipitate endogenous TEL2. Flag-HA-TTI1 was transiently transfected into 293T cells, and proteins immunoprecipitated with HA antibody-conjugated beads were separated by SDS-PAGE, transferred to PVDF, and immunoblotted with anti-TEL2 antisera or anti-Flag-HRP. (B) An antibody generated against the C terminus of TTI1 coimmunoprecipitated endogenous TTI1 and TEL2. Normal rabbit IgG was used for a control immunoprecipitation. (C) TEL2 can coimmunoprecipitate TTI1 independently of IR treatment. MYC-TTI1 (N-terminal MYC tag) and/or TEL2-FHA (C-terminal Flag-HA tag) were transiently transfected into 293T cells, and proteins were immunoprecipitated with HA antibodies and immunoblotted with anti-MYC or anti-HA antibodies. Thirty-six hours after transfection, cells were either left untreated or treated with 10 Gy of IR for 4 h prior to harvesting. (D) TTI2 interacts with both TTI1 and TEL2. FHA-TTI2 (N-terminal Flag-HA tag) was transiently transfected into 293T cells, and HA immunoprecipitates were immunoblotted with anti-TEL2 and anti-TTI1 antibodies. (E) TTI1, TEL2, and TTI2 cofractionate in a large-molecular-weight complex on Superose 6. 293 TRex cells containing a doxycycline-inducible Flag-HA-TTI2 were treated with 2 ug/mL doxycycline for 24 h prior to preparation of whole-cell extract. The molecular weight standards and the corresponding fractions they eluted off of the Superose 6 column are listed in Supplemental Table S5. (F) Fractions from extracts run on a Superose 6 column (shown in E) were used as input for immunoprecipitation with HA antibody-conjugated beads.
Figure 7.
TTI1 and TTI2 are required for maintaining TEL2 protein levels, and are critical components of the TTT complex. (A) TTI1 is required to maintain TEL2 protein levels. Whole-cell extracts from U2OS cells stably expressing control, ATR, or TTI1 shRNAs were immunoblotted with TEL2, TTI1, or VINCULIN antisera. (B) Expression of an shRNA-resistant TTI1 cDNA can restore TEL2 and PIKK protein levels. U2OS cells stably expressing a control (FF) or TTI1 shRNA #3 (targeting the 3′UTR of endogenous TTI1) were infected with an empty vector or Flag-HA-TTI1 for 2 d. Whole-cell extracts were immunoblotted with the indicated antisera. (C) Overexpression of TEL2 does not restore IR resistance to TTI1-depleted cells. U2OS cells stably expressing TTI1 shRNA #3 were infected with retroviruses expressing MSCV-HA-Flag-empty, MSCV-HA-Flag-TTI1, or MSCV-TEL2-HA-Flag, and, after 2 d, were mixed with control cells for MCA. Viability in response to 3 Gy of IR or 75 nM MMC was monitored. (D) Each subunit of the TTT complex is required for DNA damage resistance. MCA was performed using U2OS cells expressing dsRed, and the individual shRNAs are indicated as in Figure 2C. Viability in response to 3 Gy of IR or 75 nM MMC was monitored. shRNA sequences are listed in Supplemental Table S6. (E) Knockdown of TTT subunits affects ATM and ATR levels and levels of the other TTT subunits. Immunoblots corresponding to the viability assays shown in D. The lysates from cells expressing FF, ATM, ATR, TTI1 #1, and TTI1 #2 shRNAs are the same in both panels to allow for comparison between the effects of TEL2 and TTI2 depletion. (F) TTI1 is critical for interaction of the TTT complex with ATM. U2OS cells were transfected with siRNA duplexes and incubated for 72 h prior to harvesting for preparation of whole-cell extracts. Immunoprecipitations were performed using control, TTI1, or TEL2 (ProteinTech Group) antisera. TTI1, TEL2, and TTI2 pools of an equimolar mix of four siRNAs from Dharmacon were used (See Supplemental Table S7).
Similar articles
- Dissecting cellular responses to irradiation via targeted disruptions of the ATM-CHK1-PP2A circuit.
Palii SS, Cui Y, Innes CL, Paules RS. Palii SS, et al. Cell Cycle. 2013 Apr 1;12(7):1105-18. doi: 10.4161/cc.24127. Epub 2013 Mar 5. Cell Cycle. 2013. PMID: 23462183 Free PMC article. - Inositol pyrophosphates mediate the DNA-PK/ATM-p53 cell death pathway by regulating CK2 phosphorylation of Tti1/Tel2.
Rao F, Cha J, Xu J, Xu R, Vandiver MS, Tyagi R, Tokhunts R, Koldobskiy MA, Fu C, Barrow R, Wu M, Fiedler D, Barrow JC, Snyder SH. Rao F, et al. Mol Cell. 2014 Apr 10;54(1):119-132. doi: 10.1016/j.molcel.2014.02.020. Epub 2014 Mar 20. Mol Cell. 2014. PMID: 24657168 Free PMC article. - Tel2 regulates the stability of PI3K-related protein kinases.
Takai H, Wang RC, Takai KK, Yang H, de Lange T. Takai H, et al. Cell. 2007 Dec 28;131(7):1248-59. doi: 10.1016/j.cell.2007.10.052. Cell. 2007. PMID: 18160036 - The role of NBS1 in the modulation of PIKK family proteins ATM and ATR in the cellular response to DNA damage.
Zhou J, Lim CU, Li JJ, Cai L, Zhang Y. Zhou J, et al. Cancer Lett. 2006 Nov 8;243(1):9-15. doi: 10.1016/j.canlet.2006.01.026. Epub 2006 Mar 10. Cancer Lett. 2006. PMID: 16530324 Free PMC article. Review. - The role of ATM and ATR in DNA damage-induced cell cycle control.
Goodarzi AA, Block WD, Lees-Miller SP. Goodarzi AA, et al. Prog Cell Cycle Res. 2003;5:393-411. Prog Cell Cycle Res. 2003. PMID: 14593734 Review.
Cited by
- Systems genetics in the rat HXB/BXH family identifies Tti2 as a pleiotropic quantitative trait gene for adult hippocampal neurogenesis and serum glucose.
Senko AN, Overall RW, Silhavy J, Mlejnek P, Malínská H, Hüttl M, Marková I, Fabel KS, Lu L, Stuchlik A, Williams RW, Pravenec M, Kempermann G. Senko AN, et al. PLoS Genet. 2022 Apr 4;18(4):e1009638. doi: 10.1371/journal.pgen.1009638. eCollection 2022 Apr. PLoS Genet. 2022. PMID: 35377872 Free PMC article. - The C-terminal residues of Saccharomyces cerevisiae Mec1 are required for its localization, stability, and function.
DaSilva LF, Pillon S, Genereaux J, Davey MJ, Gloor GB, Karagiannis J, Brandl CJ. DaSilva LF, et al. G3 (Bethesda). 2013 Oct 3;3(10):1661-74. doi: 10.1534/g3.113.006841. G3 (Bethesda). 2013. PMID: 23934994 Free PMC article. - Making sense of cancer genomic data.
Chin L, Hahn WC, Getz G, Meyerson M. Chin L, et al. Genes Dev. 2011 Mar 15;25(6):534-55. doi: 10.1101/gad.2017311. Genes Dev. 2011. PMID: 21406553 Free PMC article. Review. - Genome-wide CRISPR screens identify novel regulators of wild-type and mutant p53 stability.
Lü Y, Cho T, Mukherjee S, Suarez CF, Gonzalez-Foutel NS, Malik A, Martinez S, Dervovic D, Oh RH, Langille E, Al-Zahrani KN, Hoeg L, Lin ZY, Tsai R, Mbamalu G, Rotter V, Ashton-Prolla P, Moffat J, Chemes LB, Gingras AC, Oren M, Durocher D, Schramek D. Lü Y, et al. Mol Syst Biol. 2024 Jun;20(6):719-740. doi: 10.1038/s44320-024-00032-x. Epub 2024 Apr 5. Mol Syst Biol. 2024. PMID: 38580884 Free PMC article. - The Connection Between Cell Fate and Telomere.
Engin AB, Engin A. Engin AB, et al. Adv Exp Med Biol. 2021;1275:71-100. doi: 10.1007/978-3-030-49844-3_3. Adv Exp Med Biol. 2021. PMID: 33539012 Review.
References
- Ahmed S, Alpi A, Hengartner MO, Gartner A 2001. C. elegans RAD-5/CLK-2 defines a new DNA damage checkpoint protein. Curr Biol 11: 1934–1944 - PubMed
- Benard C, McCright B, Zhang Y, Felkai S, Lakowski B, Hekimi S 2001. The C. elegans maternal-effect gene clk-2 is essential for embryonic development, encodes a protein homologous to yeast Tel2p and affects telomere length. Development 128: 4045–4055 - PubMed
- Collis SJ, Barber LJ, Clark AJ, Martin JS, Ward JD, Boulton SJ 2007. HCLK2 is essential for the mammalian S-phase checkpoint and impacts on Chk1 stability. Nat Cell Biol 9: 391–401 - PubMed
- Collis SJ, Ciccia A, Deans AJ, Horejsi Z, Martin JS, Maslen SL, Skehel JM, Elledge SJ, West SC, Boulton SJ 2008. FANCM and FAAP24 function in ATR-mediated checkpoint signaling independently of the Fanconi anemia core complex. Mol Cell 32: 313–324 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous