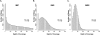Systematic comparison of three genomic enrichment methods for massively parallel DNA sequencing - PubMed (original) (raw)
Comparative Study
. 2010 Oct;20(10):1420-31.
doi: 10.1101/gr.106716.110. Epub 2010 Sep 1.
Lori L Bonnycastle, Peter S Chines, Nancy F Hansen, Natsuyo Aoyama, Amy J Swift, Hatice Ozel Abaan, Thomas J Albert; NISC Comparative Sequencing Program; Elliott H Margulies, Eric D Green, Francis S Collins, James C Mullikin, Leslie G Biesecker
Affiliations
- PMID: 20810667
- PMCID: PMC2945191
- DOI: 10.1101/gr.106716.110
Comparative Study
Systematic comparison of three genomic enrichment methods for massively parallel DNA sequencing
Jamie K Teer et al. Genome Res. 2010 Oct.
Abstract
Massively parallel DNA sequencing technologies have greatly increased our ability to generate large amounts of sequencing data at a rapid pace. Several methods have been developed to enrich for genomic regions of interest for targeted sequencing. We have compared three of these methods: Molecular Inversion Probes (MIP), Solution Hybrid Selection (SHS), and Microarray-based Genomic Selection (MGS). Using HapMap DNA samples, we compared each of these methods with respect to their ability to capture an identical set of exons and evolutionarily conserved regions associated with 528 genes (2.61 Mb). For sequence analysis, we developed and used a novel Bayesian genotype-assigning algorithm, Most Probable Genotype (MPG). All three capture methods were effective, but sensitivities (percentage of targeted bases associated with high-quality genotypes) varied for an equivalent amount of pass-filtered sequence: for example, 70% (MIP), 84% (SHS), and 91% (MGS) for 400 Mb. In contrast, all methods yielded similar accuracies of >99.84% when compared to Infinium 1M SNP BeadChip-derived genotypes and >99.998% when compared to 30-fold coverage whole-genome shotgun sequencing data. We also observed a low false-positive rate with all three methods; of the heterozygous positions identified by each of the capture methods, >99.57% agreed with 1M SNP BeadChip, and >98.840% agreed with the whole-genome shotgun data. In addition, we successfully piloted the genomic enrichment of a set of 12 pooled samples via the MGS method using molecular bar codes. We find that these three genomic enrichment methods are highly accurate and practical, with sensitivities comparable to that of 30-fold coverage whole-genome shotgun data.
Figures
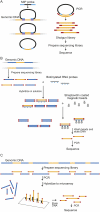
Figure 1.
Three genomic enrichment methods. (A) (MIP) Molecular Inversion Probe: 70 base probes are prepared and hybridized to genomic DNA. Capture occurs by filling in sequence between the probe-targeting arms with polymerase and then sealing the circle with ligase. Total genomic DNA is removed with nucleases. The remaining closed circles undergo shotgun library and sequencing library preparation, followed by sequencing. (B) (SHS) Solution Hybrid Selection: A sequencing library is prepared from genomic DNA. This library is hybridized to biotinylated RNA probes in solution and recovered with streptavidin beads. Eluted products are amplified prior to cluster generation and sequencing. (C) (MGS) Microarray-based Genomic Selection: A sequencing library is prepared from genomic DNA and hybridized to a capture array. Eluted products are amplified prior to cluster generation and sequencing.
Figure 2.
Depth of coverage distribution. Distributions of depth of coverage at each ROI position. Scales have been standardized for comparison purposes, and maximum coverage depth values are indicated above the arrow. (A) (MIP) Molecular Inversion Probe. (B) (SHS) Solution Hybrid Selection. (C) (MGS) Microarray-based Genomic Selection.
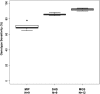
Figure 3.
Genotype sensitivity across multiple samples. Boxplots showing distribution of genotype call sensitivities across multiple samples (extended sample set) for each capture method. (N) Number of samples.
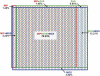
Figure 4.
ROI regions with genotype assignments. The Venn diagram of overlapping genotype coverage is area proportional. Colored rectangles identify the proportion of genotype assignments in the ROI for each method: (red) MIP; (green) SHS; (blue) MGS. Note that the greatest overlap is among all three methods, and the second greatest is between SHS and MGS. The numbers sum to 95.35% because 4.65% of the ROI was not assigned a genotype in any method.
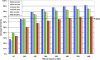
Figure 5.
Genotype sensitivity with increasing sequence data. Percentage of genotypes assigned in the ROI (same for all methods) with increasing filtered sequence data for NA18507 and NA12878. Sequence counts are based on 36 bases per read for MIP and SHS. To account for the 6-base index bar code, 42 bases were used in the sequence count calculations for MGS. The dashed arrow indicates genotype sensitivity level (67.3%) of 30-fold coverage whole-genome shotgun (WG) data for the ROI analyzed in this study.
Similar articles
- Comparison of solution-based exome capture methods for next generation sequencing.
Sulonen AM, Ellonen P, Almusa H, Lepistö M, Eldfors S, Hannula S, Miettinen T, Tyynismaa H, Salo P, Heckman C, Joensuu H, Raivio T, Suomalainen A, Saarela J. Sulonen AM, et al. Genome Biol. 2011 Sep 28;12(9):R94. doi: 10.1186/gb-2011-12-9-r94. Genome Biol. 2011. PMID: 21955854 Free PMC article. - Solution hybrid selection with ultra-long oligonucleotides for massively parallel targeted sequencing.
Gnirke A, Melnikov A, Maguire J, Rogov P, LeProust EM, Brockman W, Fennell T, Giannoukos G, Fisher S, Russ C, Gabriel S, Jaffe DB, Lander ES, Nusbaum C. Gnirke A, et al. Nat Biotechnol. 2009 Feb;27(2):182-9. doi: 10.1038/nbt.1523. Epub 2009 Feb 1. Nat Biotechnol. 2009. PMID: 19182786 Free PMC article. - Combining microarray-based genomic selection (MGS) with the Illumina Genome Analyzer platform to sequence diploid target regions.
Okou DT, Locke AE, Steinberg KM, Hagen K, Athri P, Shetty AC, Patel V, Zwick ME. Okou DT, et al. Ann Hum Genet. 2009 Sep;73(Pt 5):502-13. doi: 10.1111/j.1469-1809.2009.00530.x. Epub 2009 Jul 1. Ann Hum Genet. 2009. PMID: 19573206 Free PMC article. - Exome sequencing: the sweet spot before whole genomes.
Teer JK, Mullikin JC. Teer JK, et al. Hum Mol Genet. 2010 Oct 15;19(R2):R145-51. doi: 10.1093/hmg/ddq333. Epub 2010 Aug 12. Hum Mol Genet. 2010. PMID: 20705737 Free PMC article. Review. - Selective gene amplification for high-throughput sequencing.
Ni T, Wu H, Song S, Jelley M, Zhu J. Ni T, et al. Recent Pat DNA Gene Seq. 2009;3(1):29-38. doi: 10.2174/187221509787236183. Recent Pat DNA Gene Seq. 2009. PMID: 19149736 Review.
Cited by
- Application and comparison of large-scale solution-based DNA capture-enrichment methods on ancient DNA.
Avila-Arcos MC, Cappellini E, Romero-Navarro JA, Wales N, Moreno-Mayar JV, Rasmussen M, Fordyce SL, Montiel R, Vielle-Calzada JP, Willerslev E, Gilbert MT. Avila-Arcos MC, et al. Sci Rep. 2011;1:74. doi: 10.1038/srep00074. Epub 2011 Aug 24. Sci Rep. 2011. PMID: 22355593 Free PMC article. - Use of microarray hybrid capture and next-generation sequencing to identify the anatomy of a transgene.
Dubose AJ, Lichtenstein ST, Narisu N, Bonnycastle LL, Swift AJ, Chines PS, Collins FS. Dubose AJ, et al. Nucleic Acids Res. 2013 Apr 1;41(6):e70. doi: 10.1093/nar/gks1463. Epub 2013 Jan 11. Nucleic Acids Res. 2013. PMID: 23314155 Free PMC article. - Whole exome sequencing in the rat.
Foley JF, Phadke DP, Hardy O, Hardy S, Miller V, Madan A, Howard K, Kruse K, Lord C, Ramaiahgari S, Solomon GG, Shah RR, Pandiri AR, Herbert RA, Sills RC, Merrick BA. Foley JF, et al. BMC Genomics. 2018 Jun 20;19(1):487. doi: 10.1186/s12864-018-4858-8. BMC Genomics. 2018. PMID: 29925311 Free PMC article. - Combined alpha-delta platelet storage pool deficiency is associated with mutations in GFI1B.
Ferreira CR, Chen D, Abraham SM, Adams DR, Simon KL, Malicdan MC, Markello TC, Gunay-Aygun M, Gahl WA. Ferreira CR, et al. Mol Genet Metab. 2017 Mar;120(3):288-294. doi: 10.1016/j.ymgme.2016.12.006. Epub 2016 Dec 18. Mol Genet Metab. 2017. PMID: 28041820 Free PMC article. - An improved understanding of cancer genomics through massively parallel sequencing.
Teer JK. Teer JK. Transl Cancer Res. 2014 Jun;3(3):243-259. doi: 10.3978/j.issn.2218-676X.2014.05.05. Transl Cancer Res. 2014. PMID: 26146607 Free PMC article.
References
- Albert TJ, Molla MN, Muzny DM, Nazareth L, Wheeler D, Song X, Richmond TA, Middle CM, Rodesch MJ, Packard CJ, et al. 2007. Direct selection of human genomic loci by microarray hybridization. Nat Methods 4: 903–905 - PubMed
- Bau S, Schracke N, Kranzle M, Wu H, Stahler PF, Hoheisel JD, Beier M, Summerer D 2009. Targeted next-generation sequencing by specific capture of multiple genomic loci using low-volume microfluidic DNA arrays. Anal Bioanal Chem 393: 171–175 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous