Eaten alive: a history of macroautophagy - PubMed (original) (raw)
Review
Eaten alive: a history of macroautophagy
Zhifen Yang et al. Nat Cell Biol. 2010 Sep.
Abstract
Macroautophagy (hereafter autophagy), or 'self-eating', is a conserved cellular pathway that controls protein and organelle degradation, and has essential roles in survival, development and homeostasis. Autophagy is also integral to human health and is involved in physiology, development, lifespan and a wide range of diseases, including cancer, neurodegeneration and microbial infection. Although research on this topic began in the late 1950s, substantial progress in the molecular study of autophagy has taken place during only the past 15 years. This review traces the key findings that led to our current molecular understanding of this complex process.
Figures
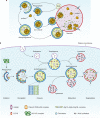
Figure 1
Schematic depiction of autophagy. (a) In yeast, both autophagy and the Cvt pathway engulf cargoes within distinct double-membrane vesicles, which are thought to originate from the phagophore assembly site (PAS). The PAS is defined as the initial site for autophagy-related (Atg) protein recruitment. The Cvt pathway is one example of selective autophagy, and the only example of a biosynthetic autophagy-related pathway. The Cvt vesicle (140–160 nm in diameter) appears to closely enwrap the specific cargo — the Cvt complex (consisting of the precursor form of aminopeptidase I — prApe1 — and the Atg19 receptor), and exclude bulk cytoplasm. The autophagosome (300–900 nm in diameter) engulfs cytoplasm, including organelles, and also the Cvt complex. The completed vesicles then fuse with the vacuole, the yeast analogue of the mammalian lysosome, and release the inner single-membrane vesicle (autophagic or Cvt body) into the lumen. Subsequent breakdown of the inner vesicles allows the maturation of prApe1 and the degradation of cytoplasm, and hence the recycling of the resulting macromolecules through vacuolar permeases. (b) Mammalian autophagy is initiated by the formation of the phagophore, followed by a series of steps, including the elongation and expansion of the phagophore, closure and completion of a double-membrane autophagosome (which surrounds a portion of the cytoplasm), autophagosome maturation through docking and fusion with an endosome (the product of fusion is known as an amphisome) and/or lysosome (the product of fusion is known as an autolysosome), breakdown and degradation of the autophagosome inner membrane and cargo through acid hydrolases inside the autolysosome, and recycling of the resulting macromolecules through permeases. So far, there is no evidence for a PAS that exists in mammalian cells, and so the mammalian phagophore could be equivalent to the yeast PAS, or derived from the PAS. The core molecular machinery is also depicted, such as the ULK1 and ULK2 complexes that are required for autophagy induction, class III PtdIns3K complexes that are involved in autophagosome formation, mammalian Atg9 (mAtg9) that potentially contributes to the delivery of membrane to the forming autophagosome and two conjugation systems, the LC3-II and Atg12–Atg5–Atg16L complex, which are proposed to function during elongation and expansion of the phagophore membrane.
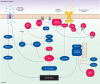
Figure 2
Signalling regulation of mammalian autophagy. In the figure, the blue components represent the factors that stimulate autophagy, whereas the red ones correspond to inhibitory factors. Autophagy is regulated by a complex signalling network of various stimulatory (blue arrows) and inhibitory (red bars) inputs. TOR plays a central role in autophagy by integrating the class I PtdIns3K signalling and amino acid-dependent signalling pathways. Activation of insulin receptors stimulates the class I PtdIns3K complex and small GTPase Ras, leading to activation of the PtdIns3K–PKB–TOR pathway. PKB phosphorylates and inhibits the tuberous sclerosis complex 1/2 (TSC1–TSC2), leading to the stabilization of Rheb GTPase, which in turn activates TOR, causing inhibition of autophagy. Amino acids inhibit the Raf-1–MEK1/2–ERK1/2 signalling cascade, leading to inhibition of autophagy. Energy depletion causes the AMP-activated protein kinase (AMPK) to be phosphorylated and activated by LKB1. AMPK phosphorylates and activates TSC1–TSC2, leading to inactivation of TOR and autophagy induction. p70S6K kinase is a substrate of TOR that may negatively feed back on TOR activity, ensuring basal levels of autophagy that are important for homeostasis. JNK1 and DAPK phosphorylate and disrupt the association of anti-apoptotic proteins, Bcl-2 and Bcl-XL, with Beclin 1, leading to the activation of the Beclin 1-associated class III PtdIns3K complex and stimulation of autophagy. Beclin 1 is shown bound to the phagophore membrane.
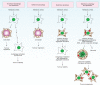
Figure 3
A model for the roles of apoptosis and autophagy in tumorigenesis. A common cellular response to metabolic stress is cell death mediated by apoptosis, which limits tumour growth. Tumours may trigger autophagy-mediated cell survival in certain metabolic-stressed tumour regions. In apoptotic-defective, metabolic-stressed tumour cells, activation of autophagy prevents death from necrosis, whereas defects in autophagy lead to accumulation of p62, damaged mitochondria, ROS and protein aggregates, resulting in genome damage and tumorigenesis. For additional information, see refs and .
Similar articles
- Chaperone-mediated autophagy: the heretofore untold story of J. Fred "Paulo" Dice. Interview by Daniel J. Klionsky.
Dice JF. Dice JF. Autophagy. 2009 Nov;5(8):1079-84. doi: 10.4161/auto.5.8.9476. Epub 2009 Nov 21. Autophagy. 2009. PMID: 19617700 - A brief history of autophagy from cell biology to physiology and disease.
Mizushima N. Mizushima N. Nat Cell Biol. 2018 May;20(5):521-527. doi: 10.1038/s41556-018-0092-5. Epub 2018 Apr 23. Nat Cell Biol. 2018. PMID: 29686264 Review. - Autophagy researchers.
Boya P, Diaz-Meco MT, Rubinsztein D, Sass M. Boya P, et al. Autophagy. 2014 Mar;10(3):393-6. doi: 10.4161/auto.27581. Autophagy. 2014. PMID: 24721973 Free PMC article. No abstract available. - From the urea cycle to autophagy: Alfred J. Meijer.
Meijer AJ. Meijer AJ. Autophagy. 2011 Aug;7(8):805-13. doi: 10.4161/auto.7.8.15192. Epub 2011 Aug 1. Autophagy. 2011. PMID: 21389787 - The significance of macroautophagy in health and disease.
Tukaj C. Tukaj C. Folia Morphol (Warsz). 2013 May;72(2):87-93. doi: 10.5603/fm.2013.0015. Folia Morphol (Warsz). 2013. PMID: 23740493 Review.
Cited by
- The ULK1 complex: sensing nutrient signals for autophagy activation.
Wong PM, Puente C, Ganley IG, Jiang X. Wong PM, et al. Autophagy. 2013 Feb 1;9(2):124-37. doi: 10.4161/auto.23323. Epub 2013 Jan 7. Autophagy. 2013. PMID: 23295650 Free PMC article. Review. - Cell loss and autophagy in the extra-adrenal chromaffin organ of Zuckerkandl are regulated by glucocorticoid signalling.
Schober A, Parlato R, Huber K, Kinscherf R, Hartleben B, Huber TB, Schütz G, Unsicker K. Schober A, et al. J Neuroendocrinol. 2013 Jan;25(1):34-47. doi: 10.1111/j.1365-2826.2012.02367.x. J Neuroendocrinol. 2013. PMID: 23078542 Free PMC article. - Elevated Mirc1/Mir17-92 cluster expression negatively regulates autophagy and CFTR (cystic fibrosis transmembrane conductance regulator) function in CF macrophages.
Tazi MF, Dakhlallah DA, Caution K, Gerber MM, Chang SW, Khalil H, Kopp BT, Ahmed AE, Krause K, Davis I, Marsh C, Lovett-Racke AE, Schlesinger LS, Cormet-Boyaka E, Amer AO. Tazi MF, et al. Autophagy. 2016 Nov;12(11):2026-2037. doi: 10.1080/15548627.2016.1217370. Autophagy. 2016. PMID: 27541364 Free PMC article. - Gefitinib induces lung cancer cell autophagy and apoptosis via blockade of the PI3K/AKT/mTOR pathway.
Zhao ZQ, Yu ZY, Li J, Ouyang XN. Zhao ZQ, et al. Oncol Lett. 2016 Jul;12(1):63-68. doi: 10.3892/ol.2016.4606. Epub 2016 May 18. Oncol Lett. 2016. PMID: 27347100 Free PMC article. - Novel Insights into the Interplay between Apoptosis and Autophagy.
Rikiishi H. Rikiishi H. Int J Cell Biol. 2012;2012:317645. doi: 10.1155/2012/317645. Epub 2012 Mar 15. Int J Cell Biol. 2012. PMID: 22496691 Free PMC article.
References
- de Duve C, Wattiaux R. Functions of lysosomes. Annu. Rev. Physiol. 1966;28:435–492. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous