Selectivity mechanism of the nuclear pore complex characterized by single cargo tracking - PubMed (original) (raw)
Selectivity mechanism of the nuclear pore complex characterized by single cargo tracking
Alan R Lowe et al. Nature. 2010.
Abstract
The nuclear pore complex (NPC) mediates all exchange between the cytoplasm and the nucleus. Small molecules can passively diffuse through the NPC, whereas larger cargos require transport receptors to translocate. How the NPC facilitates the translocation of transport receptor/cargo complexes remains unclear. To investigate this process, we tracked single protein-functionalized quantum dot cargos as they moved through human NPCs. Here we show that import proceeds by successive substeps comprising cargo capture, filtering and translocation, and release into the nucleus. Most quantum dots are rejected at one of these steps and return to the cytoplasm, including very large cargos that abort at a size-selective barrier. Cargo movement in the central channel is subdiffusive and cargos that can bind more transport receptors diffuse more freely. Without Ran GTPase, a critical regulator of transport directionality, cargos still explore the entire NPC, but have a markedly reduced probability of exit into the nucleus, suggesting that NPC entry and exit steps are not equivalent and that the pore is functionally asymmetric to importing cargos. The overall selectivity of the NPC seems to arise from the cumulative action of multiple reversible substeps and a final irreversible exit step.
Conflict of interest statement
Competing financial interests: The authors declare no competing financial interests.
Figures
Figure 1. Schematic of Experiment
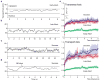
(a) Schematic of QD based cargo. The Snurportin-1 IBB/Z-domain fusion protein is coupled via a bifunctional SMCC crosslinker to the amino-PEG polymer coat of a fluorescent QD. The three helix Z-domain acts as a spacer to correctly present the IBB for biological function. Not to scale. (b) Dynamic Light Scattering size distributions of QD-IBB cargos in the presence and absence of importin-β. (c) Dwell time distribution of all QD interactions with the NPC. The time axis is truncated at 300 s. (d) Brightfield image of a nucleus with a quantum dot fluorescence image (with background subtraction applied) overlaid in red. A single quantum dot cargo at the nuclear envelope is highlighted. (e) Individual consecutive frames from a single-molecule experiment showing the arrival (first frame) from the cytoplasm and departure (final frame) of the cargo into the nucleus. The centroids determined from fitting of the PSF are overlaid as red crosses. Frame numbers are in the bottom left hand corner of each frame. Movies were captured at 40 Hz.
Figure 2. Examples of single-molecule trajectories and a functional map of the NPC interior
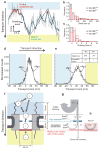
(a) Left, Schematic of the nuclear pore complex showing approximate sizes from the literature. The QD-IBB cargo is shown as a red circle to scale. Right, Three examples each of aborted and successful import trajectories (0.1 s running mean filtered) and coloured according to sub-steps: Docking events are coloured in blue, motion in the central channel in green and undocking in red. Raw, unfiltered data of cargos showing rapid cargo exit into the nucleus are provided as Supplementary Fig. S12. (b) Contour maps of the density of found cargo positions in the central channel for all successful, all aborted, early aborts, and late aborts. The contours are coloured according to the normalised density of observations, with red representing the highest density of found cargo positions and blue the lowest (See supplementary materials for details of trajectory alignment). (c–e) Positional histograms along the transport axis, for successful, early and late aborting cargos.
Figure 3. Cargo Motion in the NPC central channel
(a,b) Examples of early and late aborting cargos. Kymographs showing motion along the transport axis, X, as a function of time (grey, raw data; red, 0.1 s running mean filtered). (c) An example of a successfully importing trajectory showing early confinement at an internal constriction. (d) A second example of a successful trajectory without visible early confinement. (e) Ensemble averaged Mean Squared Displacement (MSD) plots showing anomalous sub-diffusion in the transverse and transport axes of the central channel. Fits to a power law 〈_r_2〉= Γ_t_α are shown as solid lines (Supplementary Materials). Raw frame-to-frame displacements versus channel position are provided as Supplementary Fig. S13. Errors are standard error of the mean (_n_success = 23, _n_early = 11, _n_late = 22).
Figure 4. Effect of IBB density on cargo motion, location of Ran action, and data summary
(a) Detail of a crossing event within the central channel (Grey, raw data; blue line, 0.1s running average). The cargo dwells at the cytoplasmic and nuclear faces of the channel for various durations. The crossing time is the time between leaving the cytoplasmic face and arriving at the nuclear face and vice versa. (b) Cargo dwell times with 50% or 100% IBB. The mean/median dwell time increases for the 50% labelled cargo. The distributions (p<0.05, Kolmogorov Smirnov test) and medians (_p_<0.05, two-tailed Mann-Whitney test) are significantly different (Supplementary Materials). (**c**) Crossing time distributions for cargos with 50% or 100% IBB. QB-IBB50% cargos take significantly longer to cross the channel (_p_<0.05, Mann-Whitney test). (**d**,**e**) Position histograms of early (**d**) and late (**e**) aborts with and without Ran. Inset: import statistics with and without Ran. The presence of Ran significantly increases the probability of successful import. (**f**) Results summary. Cargos arriving from the cytoplasm (white circles) may dock on the cytoplasmic filaments or directly enter the NPC. Once inside the central channel, cargos exhibit anomalous subdiffusion. There is a size selective constriction (blue) within the first 30nm of the channel. Efficient cargo exit into the nucleus requires Ran (red bar). (**g**) Probabilities of cargos being rejected versus position, highlighting the sequence of steps in cargo translocation. Many cargos interact with the cytoplasmic filaments, but most (75%) immediately rebound into the cytoplasm. The remaining 25% interact with the NPC (grey box) for longer times. Of those, 20% abort early due to a size gate and 80% reach the central channel. Once inside the channel, in the presence of Ran, 50% of cargos ultimately enter the nucleus and the remaining 50% abort. (**h**) In the absence of Ran, cargos do not enter the nucleus (>99% abort) and return to the cytoplasm.
Similar articles
- Permeating the nuclear pore complex.
Kapon R, Naim B, Zbaida D, Nevo R, Tsabari O, Reich Z. Kapon R, et al. Nucleus. 2010 Nov-Dec;1(6):475-80. doi: 10.4161/nucl.1.6.13112. Epub 2010 Jul 22. Nucleus. 2010. PMID: 21327089 Free PMC article. - Nuclear pore complex: biochemistry and biophysics of nucleocytoplasmic transport in health and disease.
Jamali T, Jamali Y, Mehrbod M, Mofrad MR. Jamali T, et al. Int Rev Cell Mol Biol. 2011;287:233-86. doi: 10.1016/B978-0-12-386043-9.00006-2. Int Rev Cell Mol Biol. 2011. PMID: 21414590 Review. - Self-regulated viscous channel in the nuclear pore complex.
Ma J, Goryaynov A, Sarma A, Yang W. Ma J, et al. Proc Natl Acad Sci U S A. 2012 May 8;109(19):7326-31. doi: 10.1073/pnas.1201724109. Epub 2012 Apr 23. Proc Natl Acad Sci U S A. 2012. PMID: 22529346 Free PMC article. - Pore relations: nuclear pore complexes and nucleocytoplasmic exchange.
Rout MP, Aitchison JD. Rout MP, et al. Essays Biochem. 2000;36:75-88. doi: 10.1042/bse0360075. Essays Biochem. 2000. PMID: 12471904 Review. - Karyopherin enrichment at the nuclear pore complex attenuates Ran permeability.
Barbato S, Kapinos LE, Rencurel C, Lim RYH. Barbato S, et al. J Cell Sci. 2020 Feb 12;133(3):jcs238121. doi: 10.1242/jcs.238121. J Cell Sci. 2020. PMID: 31932502
Cited by
- Distinct, but not completely separate spatial transport routes in the nuclear pore complex.
Yang W. Yang W. Nucleus. 2013 May-Jun;4(3):166-75. doi: 10.4161/nucl.24874. Epub 2013 May 1. Nucleus. 2013. PMID: 23669120 Free PMC article. Review. - Conformational transition of FGFR kinase activation revealed by site-specific unnatural amino acid reporter and single molecule FRET.
Perdios L, Lowe AR, Saladino G, Bunney TD, Thiyagarajan N, Alexandrov Y, Dunsby C, French PM, Chin JW, Gervasio FL, Tate EW, Katan M. Perdios L, et al. Sci Rep. 2017 Jan 3;7:39841. doi: 10.1038/srep39841. Sci Rep. 2017. PMID: 28045057 Free PMC article. - SPEED Microscopy and Its Application in Nucleocytoplasmic Transport.
Ma J, Kelich JM, Yang W. Ma J, et al. Methods Mol Biol. 2016;1411:503-18. doi: 10.1007/978-1-4939-3530-7_31. Methods Mol Biol. 2016. PMID: 27147062 Free PMC article. - Experimental approaches for addressing fundamental biological questions in living, functioning cells with single molecule precision.
Lenn T, Leake MC. Lenn T, et al. Open Biol. 2012 Jun;2(6):120090. doi: 10.1098/rsob.120090. Open Biol. 2012. PMID: 22773951 Free PMC article. Review. - Scaffold nucleoporins Nup188 and Nup192 share structural and functional properties with nuclear transport receptors.
Andersen KR, Onischenko E, Tang JH, Kumar P, Chen JZ, Ulrich A, Liphardt JT, Weis K, Schwartz TU. Andersen KR, et al. Elife. 2013 Jun 11;2:e00745. doi: 10.7554/eLife.00745. Elife. 2013. PMID: 23795296 Free PMC article.
References
- Peters R. Translocation through the nuclear pore: Kaps pave the way. Bioessays. 2009;31:466–477. - PubMed
- Stewart M. Molecular mechanism of the nuclear protein import cycle. Nat Rev Mol Cell Biol. 2007;8:195–208. - PubMed
- Weis K. The nuclear pore complex: oily spaghetti or gummy bear? Cell. 2007;130:405–407. - PubMed
- Frey S, Gorlich D. A saturated FG-repeat hydrogel can reproduce the permeability properties of nuclear pore complexes. Cell. 2007;130:512–523. - PubMed
- Frey S, Richter RP, Gorlich D. FG-rich repeats of nuclear pore proteins form a three-dimensional meshwork with hydrogel-like properties. Science. 2006;314:815–817. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- GM058065/GM/NIGMS NIH HHS/United States
- R01 GM084716/GM/NIGMS NIH HHS/United States
- R01 GM058065/GM/NIGMS NIH HHS/United States
- U54 CA143836/CA/NCI NIH HHS/United States
- R01 GM084716-02/GM/NIGMS NIH HHS/United States
- U54 CA143836-01/CA/NCI NIH HHS/United States
- R01 GM077856/GM/NIGMS NIH HHS/United States
- U54CA143836/CA/NCI NIH HHS/United States
- GM77856/GM/NIGMS NIH HHS/United States
- R01 GM077856-04/GM/NIGMS NIH HHS/United States
- R01 GM058065-13/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Miscellaneous