Genome-wide measurement of RNA secondary structure in yeast - PubMed (original) (raw)
Genome-wide measurement of RNA secondary structure in yeast
Michael Kertesz et al. Nature. 2010.
Abstract
The structures of RNA molecules are often important for their function and regulation, yet there are no experimental techniques for genome-scale measurement of RNA structure. Here we describe a novel strategy termed parallel analysis of RNA structure (PARS), which is based on deep sequencing fragments of RNAs that were treated with structure-specific enzymes, thus providing simultaneous in vitro profiling of the secondary structure of thousands of RNA species at single nucleotide resolution. We apply PARS to profile the secondary structure of the messenger RNAs (mRNAs) of the budding yeast Saccharomyces cerevisiae and obtain structural profiles for over 3,000 distinct transcripts. Analysis of these profiles reveals several RNA structural properties of yeast transcripts, including the existence of more secondary structure over coding regions compared with untranslated regions, a three-nucleotide periodicity of secondary structure across coding regions and an anti-correlation between the efficiency with which an mRNA is translated and the structure over its translation start site. PARS is readily applicable to other organisms and to profiling RNA structure in diverse conditions, thus enabling studies of the dynamics of secondary structure at a genomic scale.
Figures
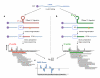
Figure 1. Measuring structural properties of RNA by deep sequencing
(a) RNA molecules are cleaved by RNase V1, which cuts 3′ of double-stranded RNA, leaving a 5′ phosphate (5′P). One such cut is illustrated by a red arrow. Following random fragmentation, V1-generated fragments are specifically captured and subjected to deep sequencing. Each aligned sequence provides structural evidence about a single base. The marked red square illustrates the evidence obtained from one mapped sequence (red). Additional evidence (gray boxes) is collected by mapping more sequences (gray horizontal bars). A large number of reads aligned to the same base indicates that the base is cleaved multiple times by RNase V1 and is thus more likely to be in double stranded conformation. (b) Same as (a), but when the RNA sample is treated with RNase S1, which cuts 3′ of single-stranded RNA. Collected reads in this case suggest that the base was unpaired in the original RNA structure. (c) By combining the data extracted from the two complementary experiments (a) and (b), we obtain a nucleotide-resolution score representing the likelihood that the inspected base was in a double- or single-stranded conformation.
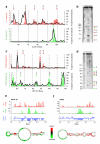
Figure 2. PARS correctly recapitulates results of RNA footprinting and known structures
(a) The PARS signal obtained for bases 50-110 of the yeast gene CCW12 using the double-stranded cutter RNase V1 (red bars) or single-stranded cutter RNase S1 (green bars) accurately matches the signals obtained by traditional footprinting of that same transcript domain (black lines). PARS signal is shown as the number of sequence reads which mapped to each nucleotide; footprinting results are obtained by semi-automated quantification of the RNase lanes shown in (b). The red arrows indicate RNase V1 cleavages and the green arrows indicate RNase S1 cleavages as shown in the gel (b). (b) Gel analysis of RNase V1 (lanes 5,6) and S1 (lanes 3,4) probing of CCW12. Additionally, RNase T1 ladder (lanes 2,8), alkaline hydrolysis (lanes 1,9), and no RNase treatment (lane 7) are shown. (c) The PARS signal obtained from bases 50-120 of the yeast gene RPL41A matches the signals obtained by traditional footprinting. (d) RNase V1 (lanes 5,6) and S1 (lanes 7,8) probing of RPL41A, RNase T1 ladder (lane 2), alkaline hydrolysis (lanes 1,9), and no RNase treatment (lane 4). (e-f) Raw number of reads obtained using RNase V1 (red bars) or RNase S1 (green bars) and the resulting PARS score (blue bars) along one inspected domain of ASH1 (e) and URE2 (f). Also shown are the known structures of the inspected domains with nucleotides color-coded according to their computed PARS score.
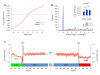
Figure 3. Functional units of the transcript are demarcated by distinct properties of RNA structure
(a) Significant correspondence between PARS and computational predictions of RNA structure. We used the Vienna package to fold the 3000 yeast mRNAs used in our analysis, and extracted the predicted double-stranded probability of each nucleotide. Shown is the average predicted double-stranded probability of each nucleotide (y-axis), where nucleotides were sorted by their PARS score (x-axis). Average and standard deviation from 1000 shuffle experiments in which a random prediction score was assigned to each probed base are shown in gray. (b) Discrete Fourier transform of average PARS score across the coding region, 3′ UTR and 5′ UTR. Inset shows PARS score obtained for each of the three positions of every codon, averaged across all codons. (c) PARS score across the 5′ UTR, the coding region, and the 3′ UTR, averaged across all transcripts used in our analysis. Transcripts were aligned by their translational start and stop sites for the left and right panel, respectively; start and stop codons are indicated by gray bars; horizontal bars denote the average PARS score per region.
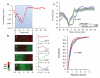
Figure 4. Structure around start codons correlates with low translational efficiency
(a) Sliding window analysis of local PARS score and ribosome density. Shown is the significance (p-value) of the anti-correlation between average PARS score along a 40bp-wide window and the reported ribosome density. (b) Left: _k_-means clustering of PARS scores across the ±40bp window surrounding the translation start site of all transcripts for which enough coverage was obtained. The average structural profile and number of member genes is shown to the right of each cluster. Right: Cumulative distribution plot of ribosome occupancy for each cluster and the associated Kolmogorov-Smirnoff test p-value between the distribution of cluster 1 and 4. (c) Tendency for less RNA structure in the first 30 bases of ORFs encoding predicted secretory proteins. While structure typically builds up immediately upon entry to the coding sequence (CDS), genes predicted to code for secretory proteins retain low structure in the first ~30 bases of the CDS, consistent with the dual function SSCR having structural features of UTR rather than CDS. Shown are the average relative PARS scores (Methods) across a 30bp sliding window for the 499 genes coding for secretory proteins (blue), the remaining 2501 genes (green) and the mean and standard deviation obtained from 1000 shuffle experiments in which sets of 499 genes were randomly selected (gray).
Comment in
- Toward global RNA structure analysis.
Mauger DM, Weeks KM. Mauger DM, et al. Nat Biotechnol. 2010 Nov;28(11):1178-9. doi: 10.1038/nbt1110-1178. Nat Biotechnol. 2010. PMID: 21057487 Free PMC article. No abstract available.
Similar articles
- RNA secondary structure profiling in zebrafish reveals unique regulatory features.
Kaushik K, Sivadas A, Vellarikkal SK, Verma A, Jayarajan R, Pandey S, Sethi T, Maiti S, Scaria V, Sivasubbu S. Kaushik K, et al. BMC Genomics. 2018 Feb 15;19(1):147. doi: 10.1186/s12864-018-4497-0. BMC Genomics. 2018. PMID: 29448945 Free PMC article. - Sequence-structure relationships in yeast mRNAs.
Chursov A, Walter MC, Schmidt T, Mironov A, Shneider A, Frishman D. Chursov A, et al. Nucleic Acids Res. 2012 Feb;40(3):956-62. doi: 10.1093/nar/gkr790. Epub 2011 Sep 27. Nucleic Acids Res. 2012. PMID: 21954438 Free PMC article. - Genome-wide probing of RNA structure reveals active unfolding of mRNA structures in vivo.
Rouskin S, Zubradt M, Washietl S, Kellis M, Weissman JS. Rouskin S, et al. Nature. 2014 Jan 30;505(7485):701-5. doi: 10.1038/nature12894. Epub 2013 Dec 15. Nature. 2014. PMID: 24336214 Free PMC article. - Programmed +1 translational frameshifting in the yeast Saccharomyces cerevisiae results from disruption of translational error correction.
Stahl G, Ben Salem S, Li Z, McCarty G, Raman A, Shah M, Farabaugh PJ. Stahl G, et al. Cold Spring Harb Symp Quant Biol. 2001;66:249-58. doi: 10.1101/sqb.2001.66.249. Cold Spring Harb Symp Quant Biol. 2001. PMID: 12762026 Review. No abstract available. - Insights into RNA structure and function from genome-wide studies.
Mortimer SA, Kidwell MA, Doudna JA. Mortimer SA, et al. Nat Rev Genet. 2014 Jul;15(7):469-79. doi: 10.1038/nrg3681. Epub 2014 May 13. Nat Rev Genet. 2014. PMID: 24821474 Review.
Cited by
- Number variation of high stability regions is correlated with gene functions.
Mao Y, Li Q, Wang W, Liang P, Tao S. Mao Y, et al. Genome Biol Evol. 2013;5(3):484-93. doi: 10.1093/gbe/evt020. Genome Biol Evol. 2013. PMID: 23407773 Free PMC article. - Identification of RNA structures and their roles in RNA functions.
Cao X, Zhang Y, Ding Y, Wan Y. Cao X, et al. Nat Rev Mol Cell Biol. 2024 Oct;25(10):784-801. doi: 10.1038/s41580-024-00748-6. Epub 2024 Jun 26. Nat Rev Mol Cell Biol. 2024. PMID: 38926530 Review. - diffBUM-HMM: a robust statistical modeling approach for detecting RNA flexibility changes in high-throughput structure probing data.
Marangio P, Law KYT, Sanguinetti G, Granneman S. Marangio P, et al. Genome Biol. 2021 May 27;22(1):165. doi: 10.1186/s13059-021-02379-y. Genome Biol. 2021. PMID: 34044851 Free PMC article. - Global identification of conserved post-transcriptional regulatory programs in trypanosomatids.
Najafabadi HS, Lu Z, MacPherson C, Mehta V, Adoue V, Pastinen T, Salavati R. Najafabadi HS, et al. Nucleic Acids Res. 2013 Oct;41(18):8591-600. doi: 10.1093/nar/gkt647. Epub 2013 Jul 22. Nucleic Acids Res. 2013. PMID: 23877242 Free PMC article. - Long non-coding RNAs in haematological malignancies.
Garitano-Trojaola A, Agirre X, Prósper F, Fortes P. Garitano-Trojaola A, et al. Int J Mol Sci. 2013 Jul 24;14(8):15386-422. doi: 10.3390/ijms140815386. Int J Mol Sci. 2013. PMID: 23887658 Free PMC article. Review.
References
- Takizawa PA, DeRisi JL, Wilhelm JE, Vale RD. Plasma membrane compartmentalization in yeast by messenger RNA transport and a septin diffusion barrier. Science. 2000;290:341–4. - PubMed
- Tucker BJ, Breaker RR. Riboswitches as versatile gene control elements. Curr Opin Struct Biol. 2005;15:342–8. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous