Characterizing the metabolism of Dehalococcoides with a constraint-based model - PubMed (original) (raw)
Characterizing the metabolism of Dehalococcoides with a constraint-based model
M Ahsanul Islam et al. PLoS Comput Biol. 2010.
Abstract
Dehalococcoides strains respire a wide variety of chloro-organic compounds and are important for the bioremediation of toxic, persistent, carcinogenic, and ubiquitous ground water pollutants. In order to better understand metabolism and optimize their application, we have developed a pan-genome-scale metabolic network and constraint-based metabolic model of Dehalococcoides. The pan-genome was constructed from publicly available complete genome sequences of Dehalococcoides sp. strain CBDB1, strain 195, strain BAV1, and strain VS. We found that Dehalococcoides pan-genome consisted of 1118 core genes (shared by all), 457 dispensable genes (shared by some), and 486 unique genes (found in only one genome). The model included 549 metabolic genes that encoded 356 proteins catalyzing 497 gene-associated model reactions. Of these 497 reactions, 477 were associated with core metabolic genes, 18 with dispensable genes, and 2 with unique genes. This study, in addition to analyzing the metabolism of an environmentally important phylogenetic group on a pan-genome scale, provides valuable insights into Dehalococcoides metabolic limitations, low growth yields, and energy conservation. The model also provides a framework to anchor and compare disparate experimental data, as well as to give insights on the physiological impact of "incomplete" pathways, such as the TCA-cycle, CO(2) fixation, and cobalamin biosynthesis pathways. The model, referred to as iAI549, highlights the specialized and highly conserved nature of Dehalococcoides metabolism, and suggests that evolution of Dehalococcoides species is driven by the electron acceptor availability.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
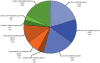
Figure 1. Composition of the Dehalococcoides pan-genome.
Core, dispensable, and unique genomes are represented by blue, green, and orange, respectively. Genes in these genomes are also categorized as metabolic (spotted pattern), non-metabolic (plain), and hypothetical (grid pattern) on the basis of various bioinformatic analyses (see text for details).
Figure 2. Distribution of dispensable and unique metabolic genes in different Dehalococcoides strains.
Colors are assigned to further categorize the genes according to their function identified from annotation and verified by different bioinformatic analyses. Each color except black signifies the presence of a corresponding metabolic gene while black indicates the absence of the corresponding gene. Genes belonging to amino acid metabolism, lipid metabolism and nucleotide metabolism are small in number; hence, included in ‘other’ category. This heat map essentially describes the differences among Dehalococcoides strains from the context of metabolic genes.
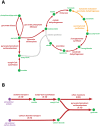
Figure 3. The reconstructed TCA-cycle and CO2 fixation pathway of Dehalococcoides.
The arrows show the directionality of the reactions. (A) Grey: citrate synthase gene currently not identified in _i_AI549, but the pathway is suggested to be present by Tang et al. ; Orange: pathways for which homologous putative genes (∼30% amino acid sequence identity) were tentatively identified in Dehalococcoides, but are suggested to be absent by Tang et al. ; Red: pathways for which putative genes are confirmed to be present by both _i_AI549 and Tang et al. . In all cases, the TCA-cycle of Dehalococcoides is not closed which explains their inability to use acetate as an energy source. (B) Dehalococcoides' requirement of CO2 in addition to acetate for their in silico growth. The numbers are flux values in mmol.gDCW−1.h−1. During pyruvate synthesis, Dehalococcoides require 67% carbon (molar basis) from acetate and 33% (molar basis) from CO2. Thus, Dehalococcoides fix carbon via the pyruvate-ferredoxin oxidoreductase or pyruvate synthase (POR) pathway.
Figure 4. Analysis of the citrate synthase (CS) reaction on Dehalococcoides growth.
(A) In the absence of the CS reaction, the TCA-cycle operates reductively via succinyl-CoA synthetase and 2-oxoglutarate synthase for producing biomass precursors for Dehalococcoides to grow. (B) The oxidative TCA-cycle operates when the CS reaction is present, but succinyl-CoA synthetase and 2-oxoglutarate synthase are absent, as suggested by the carbon labeling experiments . However, Dehalococcoides growth remains almost unchanged with and without the CS reaction (0.0137 h−1 vs. 0.014 h−1) as represented by the flux values obtained from the growth simulations of _i_AI549.
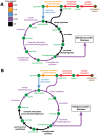
Figure 5. Reconstructed cobalamin biosynthesis pathway of Dehalococcoides.
Dashed orange lines indicate cell membrane, grey lines indicate missing pathways, and red lines indicate existing pathways, putative genes of which are identified in Dehalococcoides during the reconstruction of _i_AI549. The arrows are denoting the directionality of the reactions. Since the genomes encode a putative cobalamin transporter, Dehalococcoides may salvage vitamin B12 either in the form of cobinamide or cobalamin from the environment as indicated by ‘cobinamide transport’ and ‘cobalamin transport’ reactions in the figure. The adenosylcobalamin, which is the end product of the entire pathway, is a biomass constituent and is assumed to take part in Dehalococcoides cell formation.
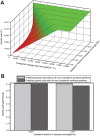
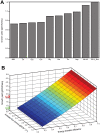
Figure 6. Influence of cobalamin on the growth rate and yield of Dehalococcoides.
(A) Growth rate of Dehalococcoides is simulated as a function of both cobalamin salvage rate and cobalamin fraction in the biomass equation. It shows the role of cobalamin in limiting the growth rate of Dehalococcoides. Clearly, the cobalamin uptake or salvage rate at which Dehalococcoides growth is limiting increases with the increase of cobalamin fraction in the biomass. (B) The cost of de novo cobalamin synthesis in terms of Dehalococcoides growth yield is compared (see text for details). The predicted yield of Dehalococcoides with and without the de novo cobalamin synthesis pathway remains almost identical for the reported maximum cobalamin fraction in the biomass. However, the predicted yield decreased only by 4% (from 0.72 gDCW/eeq to 0.69 gDCW/eeq) with 10 fold increase of cobalamin fraction in the biomass indicating the low cost of de novo cobalamin synthesis.
Figure 7. Effect of carbon and energy sources on the growth yield of Dehalococcoides.
(A) The experimental growth yield of Dehalococcoides in the minimal medium (0.69 gDCW/eeq) is compared with increased growth yields achieved by allowing unlimited fluxes of all amino acids at a H2 flux of 10 mmol.gDCW−1.h−1 (corresponding to the experimental dechlorination rate), as well as doubling the H2 flux (20 mmol.gDCW−1.h−1). It shows that unlimited flux of amino acids (carbon source) increased the in silico growth yield of Dehalococcoides by 55%, whereas doubling the H2 flux (electron donor or energy source) alone enhanced the yield by 65%. (B) Analysis of the energy limited growth of Dehalococcoides. Since the growth yield of Dehalococcoides varies linearly with the energy transfer efficiency, their yield can be improved by increasing the flux of their energy source or electron donor to generate more ATP per electron. However, the variation in acetate fluxes has no effect on growth yields. Red and green arrows show growth yields and corresponding efficiencies for Dehalococcoides growth in mixed and pure cultures, respectively. ‘MM’ = minimal medium; ‘Tyr’ = tyrosine; ‘Glu’ = glutamate; ‘Gln’ = glutamine; ‘Gly’ = glycine; ‘Ala’ = alanine; ‘Thr’ = threonine; ‘Asp’ = aspartate; ‘All AA’ = all amino acids; ‘2X H2 flux’ = 20 mmol H2.gDCW−1.h−1.
Similar articles
- New insights into Dehalococcoides mccartyi metabolism from a reconstructed metabolic network-based systems-level analysis of D. mccartyi transcriptomes.
Islam MA, Waller AS, Hug LA, Provart NJ, Edwards EA, Mahadevan R. Islam MA, et al. PLoS One. 2014 Apr 14;9(4):e94808. doi: 10.1371/journal.pone.0094808. eCollection 2014. PLoS One. 2014. PMID: 24733489 Free PMC article. - Genome sequence determination and metagenomic characterization of a Dehalococcoides mixed culture grown on cis-1,2-dichloroethene.
Yohda M, Yagi O, Takechi A, Kitajima M, Matsuda H, Miyamura N, Aizawa T, Nakajima M, Sunairi M, Daiba A, Miyajima T, Teruya M, Teruya K, Shiroma A, Shimoji M, Tamotsu H, Juan A, Nakano K, Aoyama M, Terabayashi Y, Satou K, Hirano T. Yohda M, et al. J Biosci Bioeng. 2015 Jul;120(1):69-77. doi: 10.1016/j.jbiosc.2014.12.001. Epub 2015 Jan 8. J Biosci Bioeng. 2015. PMID: 25579666 - Genome sequence of the chlorinated compound-respiring bacterium Dehalococcoides species strain CBDB1.
Kube M, Beck A, Zinder SH, Kuhl H, Reinhardt R, Adrian L. Kube M, et al. Nat Biotechnol. 2005 Oct;23(10):1269-73. doi: 10.1038/nbt1131. Epub 2005 Aug 21. Nat Biotechnol. 2005. PMID: 16116419 - The little bacteria that can - diversity, genomics and ecophysiology of 'Dehalococcoides' spp. in contaminated environments.
Taş N, van Eekert MH, de Vos WM, Smidt H. Taş N, et al. Microb Biotechnol. 2010 Jul;3(4):389-402. doi: 10.1111/j.1751-7915.2009.00147.x. Epub 2009 Sep 4. Microb Biotechnol. 2010. PMID: 21255338 Free PMC article. Review. - Evidence for nitrogen fixation by "Dehalococcoides ethenogenes" strain 195.
Lee PK, He J, Zinder SH, Alvarez-Cohen L. Lee PK, et al. Appl Environ Microbiol. 2009 Dec;75(23):7551-5. doi: 10.1128/AEM.01886-09. Epub 2009 Oct 9. Appl Environ Microbiol. 2009. PMID: 19820162 Free PMC article. Review.
Cited by
- The restricted metabolism of the obligate organohalide respiring bacterium Dehalobacter restrictus: lessons from tiered functional genomics.
Rupakula A, Kruse T, Boeren S, Holliger C, Smidt H, Maillard J. Rupakula A, et al. Philos Trans R Soc Lond B Biol Sci. 2013 Mar 11;368(1616):20120325. doi: 10.1098/rstb.2012.0325. Print 2013 Apr 19. Philos Trans R Soc Lond B Biol Sci. 2013. PMID: 23479754 Free PMC article. - Metabolic models and gene essentiality data reveal essential and conserved metabolism in prokaryotes.
Xavier JC, Patil KR, Rocha I. Xavier JC, et al. PLoS Comput Biol. 2018 Nov 16;14(11):e1006556. doi: 10.1371/journal.pcbi.1006556. eCollection 2018 Nov. PLoS Comput Biol. 2018. PMID: 30444863 Free PMC article. - Metagenomic Analysis Reveals Microbial Interactions at the Biocathode of a Bioelectrochemical System Capable of Simultaneous Trichloroethylene and Cr(VI) Reduction.
Matturro B, Zepilli M, Lai A, Majone M, Rossetti S. Matturro B, et al. Front Microbiol. 2021 Sep 30;12:747670. doi: 10.3389/fmicb.2021.747670. eCollection 2021. Front Microbiol. 2021. PMID: 34659183 Free PMC article. - Effects of Arsenic on Trichloroethene-Dechlorination Activities of Dehalococcoides mccartyi 195.
Gushgari-Doyle S, Alvarez-Cohen L. Gushgari-Doyle S, et al. Environ Sci Technol. 2020 Jan 21;54(2):1276-1285. doi: 10.1021/acs.est.9b06527. Epub 2020 Jan 8. Environ Sci Technol. 2020. PMID: 31913608 Free PMC article. - PSAMM: A Portable System for the Analysis of Metabolic Models.
Steffensen JL, Dufault-Thompson K, Zhang Y. Steffensen JL, et al. PLoS Comput Biol. 2016 Feb 1;12(2):e1004732. doi: 10.1371/journal.pcbi.1004732. eCollection 2016 Feb. PLoS Comput Biol. 2016. PMID: 26828591 Free PMC article.
References
- Medini D, Serruto D, Parkhill J, Relman DA, Donati C, et al. Microbiology in the post-genomic era. Nat Rev Microbiol. 2008;6:419–430. - PubMed
- Reed JL, Famili I, Thiele I, Palsson BO. Towards multidimensional genome annotation. Nature Reviews: Genetics. 2006;7:130–141. - PubMed
- Young JD, Henne KL, Morgan JA, Konopka AE, Ramkrishna D. Integrating cybernetic modeling with pathway analysis provides a dynamic, systems-level description of metabolic control. Biotechnol Bioeng. 2008;100:542–559. - PubMed
- Becker SA, Feist AM, Mo ML, Hannum G, Palsson BO, et al. Quantitative prediction of cellular metabolism with constraint-based models: the COBRA Toolbox. Nat Protoc. 2007;2:727–738. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases