RNA helicases at work: binding and rearranging - PubMed (original) (raw)
Review
RNA helicases at work: binding and rearranging
Eckhard Jankowsky. Trends Biochem Sci. 2011 Jan.
Abstract
RNA helicases are ubiquitous, highly conserved enzymes that participate in nearly all aspects of RNA metabolism. These proteins bind or remodel RNA or RNA-protein complexes in an ATP-dependent fashion. How RNA helicases physically perform their cellular tasks has been a longstanding question, but in recent years, intriguing models have started to link structure, mechanism and biological function for some RNA helicases. This review outlines our current view on major structural and mechanistic themes of RNA helicase function, and on emerging physical models for cellular roles of these enzymes.
Copyright © 2010 Elsevier Ltd. All rights reserved.
Figures
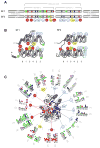
Figure 1. SF1 and SF2 helicase families
Unrooted cladogram showing the families of the SF1 (right), and the SF2 (left) according to ref. . Branch lengths are not to scale. The oval indicates significant uncertainty in cladogram topology in this region. Boldfaced names show families harboring RNA helicases (non-standard abbreviations: T1R – type 1 restriction enzymes, RHA –RNA helicase A).
Figure 2. The helicase core of SF1 and SF2 proteins
(A) Characteristic sequence motifs of SF1 and SF2 proteins in the helicase core . The motifs are colored according to their predominant biochemical function: red, ATP binding and hydrolysis; yellow, coordination between nucleic acid and NTP binding sites; blue, nucleic acid binding. Motif Ib (asterisk) is not present in all SF1 and 2 families. Green circles with asterisks designate insertions of additional domains. Distance between the conserved motifs is not to scale. (B) Location of the helicase motifs in the helicase core fold (arrows: β-strands, cylinders: α-helices). Numbers under the diagrams show connectivity of the β-strands of the first RecA-like domain. Helicase motifs are marked by circles (coloring and numbering as in panel A). Domain insertions are marked by green circles with asterisks. The rightmost β-strand and α-helix in the SF2 (blue) is not present in all SF2 families . (C) Location of the characteristic motifs in three-dimensional structure of the helicase core, as represented by SF2 DEAD-box helicase Vasa . The bound ATP analog is colored magenta, the RNA wheat. Helicase motifs are colored as in panel A. Corresponding sequence logos indicate conservation within the helicase motifs in SF1 (1) and SF2 (2). Colors mark properties of the amino acids as: green - polar, blue - basic, red – acidic, and black -hydrophobic. Circles under the letters are visual guides.
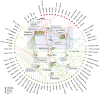
Figure 3. Cellular roles of eukaryotic RNA helicases
Selected, basic processes of eukaryotic RNA metabolism are represented by the white circles, as indicated by the callouts (NMD: nonsense mediated decay). The grey lines mark connections between processes. The colored circles represent the number of individual RNA helicases involved in a given process. RNA helicases (yeast and human orthologs) are grouped and color-coded according to their families (see legend at left lower corner). Connectors indicate involvement in one or more processes of RNA metabolism. Clear assignment of Suv3 to either SF is not possible, even though the protein is highly conserved throughout evolution . Circles with bold lines emphasize the three RNA helicases (Prp22p, Prp43p, eIF4A-III) for which specific binding site information is available.
Figure 4. Cellular functions of the DEAH/RHA helicase Prp22p and the DEAD-box protein eIF4A-III
(A) Prp22p promotes mRNA release from the spliceosome . The mRNA exons are marked red; U5snRNP aids in configuring the active site for the second splicing step. Prp22p binds the intron close to the 3' splice site. Upon exon ligation, Prp22p changes it position and binds 3' to the exon junction. Subsequently, Prp22p hydrolyzes ATP and moves towards the 5' end of the spliced mRNA, thereby removing the bound spliceosome. The figure is adapted from ref. (B) eIF4A-III acts as RNA adaptor for the other components of the EJC. The EJC is assembled during mRNA splicing and core components remain stably bound to eIF4A-III. Associated components dissociate at various stages of mRNA metabolism and further associated proteins bind throughout the travel of the EJC bound mRNA . The number of associated components shown is not representative. (C) Crystal structure of the core EJC bound to RNA . Proteins are labeled and the bound RNA is shown in orange.
Figure I
Schematic view of the main steps of translocation-based duplex unwinding. Lines represent RNA strands, the oval marks the helicase and the black rectangle indicates the ATP. Only a monomeric enzyme is displayed, but canonical helicases have also been shown to function as oligomers . Only selected, main intermediates are shown.
Figure I
Schematic view of the steps of unwinding by local strand separation. Lines represent RNA strands, the ovals mark the helicase and the small rectangles indicate the ATP/ADP. The different colors of the helicase protomers emphasize their distinct roles in the unwinding process. The asterisk after step 7 highlights the transient nature of the RNA species with a partially opened helix. Adapted, with permission, from ref. 40)
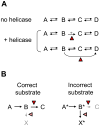
Figure I
Schemes for RNA helicase functions in the cell. (A) A series of inherently reversible reactions. The red triangle marks the helicase. (B) Proofreading by RNA helicases. The left panel shows processing of the correct substrate, the right panel processing of the incorrect substrate. The triangles mark possible action points of the helicase.
Similar articles
- Human DExD/H RNA helicases: emerging roles in stress survival regulation.
Shih JW, Lee YH. Shih JW, et al. Clin Chim Acta. 2014 Sep 25;436:45-58. doi: 10.1016/j.cca.2014.05.003. Epub 2014 May 13. Clin Chim Acta. 2014. PMID: 24835919 - Cellular functions of eukaryotic RNA helicases and their links to human diseases.
Bohnsack KE, Yi S, Venus S, Jankowsky E, Bohnsack MT. Bohnsack KE, et al. Nat Rev Mol Cell Biol. 2023 Oct;24(10):749-769. doi: 10.1038/s41580-023-00628-5. Epub 2023 Jul 20. Nat Rev Mol Cell Biol. 2023. PMID: 37474727 Review. - RNA remodeling by hexameric RNA helicases.
Rabhi M, Tuma R, Boudvillain M. Rabhi M, et al. RNA Biol. 2010 Nov-Dec;7(6):655-66. doi: 10.4161/rna.7.6.13570. Epub 2010 Nov 1. RNA Biol. 2010. PMID: 21045542 Review. - DExD/H box RNA helicases: from generic motors to specific dissociation functions.
Tanner NK, Linder P. Tanner NK, et al. Mol Cell. 2001 Aug;8(2):251-62. doi: 10.1016/s1097-2765(01)00329-x. Mol Cell. 2001. PMID: 11545728 Review. - RNA helicases: diverse roles in prokaryotic response to abiotic stress.
Owttrim GW. Owttrim GW. RNA Biol. 2013 Jan;10(1):96-110. doi: 10.4161/rna.22638. Epub 2012 Oct 23. RNA Biol. 2013. PMID: 23093803 Free PMC article. Review.
Cited by
- Steady-state NTPase activity of Dengue virus NS3: number of catalytic sites, nucleotide specificity and activation by ssRNA.
Incicco JJ, Gebhard LG, González-Lebrero RM, Gamarnik AV, Kaufman SB. Incicco JJ, et al. PLoS One. 2013;8(3):e58508. doi: 10.1371/journal.pone.0058508. Epub 2013 Mar 19. PLoS One. 2013. PMID: 23526990 Free PMC article. - In vitro studies provide insight into effects of Dicer-2 helicase mutations in Drosophila melanogaster.
Donelick HM, Talide L, Bellet M, Aruscavage PJ, Lauret E, Aguiar ERGR, Marques JT, Meignin C, Bass BL. Donelick HM, et al. RNA. 2020 Dec;26(12):1847-1861. doi: 10.1261/rna.077289.120. Epub 2020 Aug 25. RNA. 2020. PMID: 32843367 Free PMC article. - Human CWC22 escorts the helicase eIF4AIII to spliceosomes and promotes exon junction complex assembly.
Barbosa I, Haque N, Fiorini F, Barrandon C, Tomasetto C, Blanchette M, Le Hir H. Barbosa I, et al. Nat Struct Mol Biol. 2012 Oct;19(10):983-90. doi: 10.1038/nsmb.2380. Epub 2012 Sep 9. Nat Struct Mol Biol. 2012. PMID: 22961380 - DEAH-RHA helicase•Znf cofactor systems in kinetoplastid RNA editing and evolutionarily distant RNA processes.
Cruz-Reyes J, Mooers BH, Abu-Adas Z, Kumar V, Gulati S. Cruz-Reyes J, et al. RNA Dis. 2016;3(2):e1336. doi: 10.14800/rd.1336. Epub 2016 Jun 6. RNA Dis. 2016. PMID: 27540585 Free PMC article. - Measuring helicase inhibition of the DEAD-box protein Dbp2 by Yra1.
Ma WK, Tran EJ. Ma WK, et al. Methods Mol Biol. 2015;1259:183-97. doi: 10.1007/978-1-4939-2214-7_12. Methods Mol Biol. 2015. PMID: 25579587 Free PMC article.
References
- Tanner NK, Linder P. DExD/H box RNA helicases. From generic motors to specific dissociation functions. Mol Cell. 2001;8:251–261. - PubMed
- Abdelhaleem M. Do human RNA helicases have a role in cancer? Biochim Biophys Acta. 2004;1704:37–46. - PubMed
- Singleton MR, et al. Structure and mechanism of helicases and nucleic acid translocases. Ann Rev Biochem. 2007;76:23–50. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources