A genetic system to assess in vivo the functions of histones and histone modifications in higher eukaryotes - PubMed (original) (raw)
A genetic system to assess in vivo the functions of histones and histone modifications in higher eukaryotes
Ufuk Günesdogan et al. EMBO Rep. 2010 Oct.
Abstract
Despite the fundamental role of canonical histones in nucleosome structure, there is no experimental system for higher eukaryotes in which basic questions about histone function can be directly addressed. We developed a new genetic tool for Drosophila melanogaster in which the canonical histone complement can be replaced with multiple copies of experimentally modified histone transgenes. This new histone-replacement system provides a well-defined and direct cellular assay system for histone function with which to critically test models in chromatin biology dealing with chromatin assembly, variant histone functions and the biological significance of distinct histone modifications in a multicellular organism.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures
Figure 1
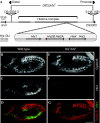
Df(2L)His C deletes all canonical histone genes. (A) Schematic representation of the canonical histone gene organization in Drosophila melanogaster. A total of 23 histone gene repeat units, each containing a single His1, His2B, His2A, His4 and His3 gene (His-GU), are clustered in the histone complex. The deletion of the histone complex includes the region between the distal rearrangement screen element CB-5033-3 and the proximal rearrangement screen element 5-HA-1581. Neighbouring genes are nrv3 and CG3305. (B–D) Wild-type histone H1 (His1) expression in S-phase 15 (S15). Cyclin B labels cells in G214; cells in early S15 show low or absent Cyclin B staining (B). S15 cells expressed His1 mRNA as detected by in situ hybridization (C). Expression was absent from G2 cells (D, Cyclin B red, His1 green). (E–G) Corresponding staining showed that His1 expression was undetectable in homozygous His C mutant embryos (His C /His C). Wild type refers to internal control. Scale bar, 100 μm; anterior, left. mRNA, messenger RNA.
Figure 2
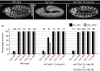
Rescue of His C by 12 His-GUs. (A–C) Representative embryos from time-matched collections 4.5–5 h after egg laying stained for Cyclin B. Wild-type embryos undergo M15 in the dorsal epidermis (A), whereas His C mutant embryos are blocked with high levels of Cyclin B before M15 (B). In the presence of 12 transgene-based His-GUs, His C mutant embryos display a wild-type M15 pattern (C); wild type refers to _w_− control. (D) The percentage of embryos that completed S15 (S15 exit) or progressed into M15 (M15 entry) in the dorsal epidermis was determined. Embryonic collections were time matched for the indicated time interval after egg laying. The left set of columns are wild type. The middle display His C homozygotes rescued by 12 His-GUs and the right display embryos from the same collection, which were not homozygous His C mutant. These embryos acted as internal controls to ensure reproducible timing of the collections. For details on classification of embryos see supplementary Fig S5 online. Scale bar, 100 μm (A–C); anterior left (A–C), His-GU; histone gene repeat unit; M15, mitosis 15; n, number of embryos.
Similar articles
- Histone supply regulates S phase timing and cell cycle progression.
Günesdogan U, Jäckle H, Herzig A. Günesdogan U, et al. Elife. 2014 Sep 9;3:e02443. doi: 10.7554/eLife.02443. Elife. 2014. PMID: 25205668 Free PMC article. - "Direct" and "Indirect" Effects of Histone Modifications: Modulation of Sterical Bulk as a Novel Source of Functionality.
Krajewski WA. Krajewski WA. Bioessays. 2020 Jan;42(1):e1900136. doi: 10.1002/bies.201900136. Epub 2019 Dec 5. Bioessays. 2020. PMID: 31805213 Review. - Transcriptional and developmental functions of the H3.3 histone variant in Drosophila.
Sakai A, Schwartz BE, Goldstein S, Ahmad K. Sakai A, et al. Curr Biol. 2009 Nov 17;19(21):1816-20. doi: 10.1016/j.cub.2009.09.021. Epub 2009 Sep 24. Curr Biol. 2009. PMID: 19781938 Free PMC article. - Histone H1, the forgotten histone.
Brockers K, Schneider R. Brockers K, et al. Epigenomics. 2019 Feb;11(4):363-366. doi: 10.2217/epi-2019-0018. Epub 2019 Feb 22. Epigenomics. 2019. PMID: 30793938 No abstract available. - Histone Post-Translational Modifications and Nucleosome Organisation in Transcriptional Regulation: Some Open Questions.
Castillo J, López-Rodas G, Franco L. Castillo J, et al. Adv Exp Med Biol. 2017;966:65-92. doi: 10.1007/5584_2017_58. Adv Exp Med Biol. 2017. PMID: 28639249 Review.
Cited by
- Dosage compensation in Drosophila.
Lucchesi JC, Kuroda MI. Lucchesi JC, et al. Cold Spring Harb Perspect Biol. 2015 May 1;7(5):a019398. doi: 10.1101/cshperspect.a019398. Cold Spring Harb Perspect Biol. 2015. PMID: 25934013 Free PMC article. Review. - Nuclear position and local acetyl-CoA production regulate chromatin state.
Willnow P, Teleman AA. Willnow P, et al. Nature. 2024 Jun;630(8016):466-474. doi: 10.1038/s41586-024-07471-4. Epub 2024 Jun 5. Nature. 2024. PMID: 38839952 Free PMC article. - Mutations that prevent or mimic persistent post-translational modifications of the histone H3 globular domain cause lethality and growth defects in Drosophila.
Graves HK, Wang P, Lagarde M, Chen Z, Tyler JK. Graves HK, et al. Epigenetics Chromatin. 2016 Feb 29;9:9. doi: 10.1186/s13072-016-0059-3. eCollection 2016. Epigenetics Chromatin. 2016. PMID: 26933451 Free PMC article. - The correlation between histone modifications and gene expression.
Dong X, Weng Z. Dong X, et al. Epigenomics. 2013 Apr;5(2):113-6. doi: 10.2217/epi.13.13. Epigenomics. 2013. PMID: 23566087 Free PMC article. No abstract available. - The Evolution of Epigenetics: From Prokaryotes to Humans and Its Biological Consequences.
Willbanks A, Leary M, Greenshields M, Tyminski C, Heerboth S, Lapinska K, Haskins K, Sarkar S. Willbanks A, et al. Genet Epigenet. 2016 Aug 3;8:25-36. doi: 10.4137/GEG.S31863. eCollection 2016. Genet Epigenet. 2016. PMID: 27512339 Free PMC article. Review.
References
- Ashburner M (1998) Drosophila: A Laboratory Handbook, 2nd edn, Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press
- Corpet A, Almouzni G (2009) Making copies of chromatin: the challenge of nucleosomal organization and epigenetic information. Trends Cell Biol 19: 29–41 - PubMed
- Henikoff S, Ahmad K (2005) Assembly of variant histones into chromatin. Annu Rev Cell Dev Biol 21: 133–153 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases