An unusual hydrophobic core confers extreme flexibility to HEAT repeat proteins - PubMed (original) (raw)
An unusual hydrophobic core confers extreme flexibility to HEAT repeat proteins
Christian Kappel et al. Biophys J. 2010.
Abstract
Alpha-solenoid proteins are suggested to constitute highly flexible macromolecules, whose structural variability and large surface area is instrumental in many important protein-protein binding processes. By equilibrium and nonequilibrium molecular dynamics simulations, we show that importin-beta, an archetypical alpha-solenoid, displays unprecedentedly large and fully reversible elasticity. Our stretching molecular dynamics simulations reveal full elasticity over up to twofold end-to-end extensions compared to its bound state. Despite the absence of any long-range intramolecular contacts, the protein can return to its equilibrium structure to within 3 A backbone RMSD after the release of mechanical stress. We find that this extreme degree of flexibility is based on an unusually flexible hydrophobic core that differs substantially from that of structurally similar but more rigid globular proteins. In that respect, the core of importin-beta resembles molten globules. The elastic behavior is dominated by nonpolar interactions between HEAT repeats, combined with conformational entropic effects. Our results suggest that alpha-solenoid structures such as importin-beta may bridge the molecular gap between completely structured and intrinsically disordered proteins.
Copyright 2010 Biophysical Society. Published by Elsevier Inc. All rights reserved.
Figures
Figure 1
Snapshots during stretching of yImp_β_. Different colors represent different HEAT repeats. The C_α_ atom of the N-terminus was kept fixed (red circle) while a moving harmonic potential was applied to the C_α_ atom of the C terminus (red arrow). (Right) Numbers denote the end-to-end distance of the protein.
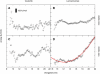
Figure 2
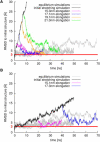
Backbone RMSD, with respect to the initial structure of yImp_β_ during stretching (black lines) and subsequent release (magenta, blue, orange, and green lines). For comparison, gray lines display data from equilibrium simulations. (Red horizontal line) RMSD value of 3 Å. (A) Stretching at 1 m/s and subsequent release at different elongations. (B) Stretching at 0.1 m/s and subsequent release at different elongations.
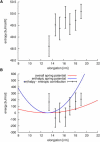
Figure 3
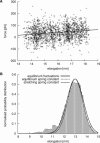
Determining the spring constant of yImp_β_. (A) Typical force curve from a slow stretching simulation (crosses) and linear fit to the data (solid line). (B) Equilibrium fluctuation of elongations of yImp_β_ (shaded bars) and derivation of the spring constant. (Broken line) Boltzmann distribution of fluctuations according to the spring constant determined from equilibrium simulations (see text). For comparison, a Boltzmann distribution according to the spring constant determined in the stretching simulations is shown (solid line).
Figure 4
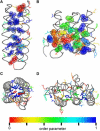
Comparison of the flexibility of hydrophobic side chains of Rop and yImp_β_. (A and C) Side and top view of Rop. (B and D) Side and top view of HEAT repeats 4–6 from yImp_β_. (Gray tubes) The protein backbone. Hydrophobic residues belonging to the respective hydrophobic core are shown as van der Waals spheres (A and B) or sticks (C and D), and as lines otherwise. The coloring reflects dihedral order parameters _S_2D (32). Values near 1 indicate rigid side chains (blue); low values reveal increased rotameric flexibility (red).
Figure 5
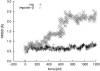
RMSD of Rop (shaded) and HEAT repeats 11 and 12 from yImp_β_ (solid) during stretching in force-probe MD simulations. For Rop, a moving harmonic potential was applied to the center-of-mass of both chains. For yImp_β_, HEAT repeats 11–13 were simulated without connecting loops between HEAT repeats. A moving harmonic potential was applied to the centers-of-mass of HEAT repeats 11 and 13. In both cases, the harmonic potentials were moved perpendicular to the main longitudinal axis. The x axis shows the sum of the applied forces on both chains and HEAT repeats, respectively.
Figure 6
Dependence of enthalpic interactions in yImp_β_ on molecular elongation. (A) Intra-HEAT-repeat Coulombic interactions. (B) Intra-HEAT-repeat Lennard-Jones interactions. (C) Inter-HEAT-repeat Coulombic interactions. (D) Inter-HEAT-repeat Lennard-Jones interactions and harmonic fit (red line). (Black bar) Energy interval of 100 kJ/mol.
Figure 7
(A) Entropy estimation for different elongations of yImp_β_. (B) Spring potential derived from purely enthalpic terms (blue line), and spring potential observed in equilibrium and stretching simulations (red line). The discrepancy between the two potentials is resolved when the entropic contribution to the free energy is subtracted from the purely enthalpic potential (black bars).
Similar articles
- Quantitative structural analysis of importin-β flexibility: paradigm for solenoid protein structures.
Forwood JK, Lange A, Zachariae U, Marfori M, Preast C, Grubmüller H, Stewart M, Corbett AH, Kobe B. Forwood JK, et al. Structure. 2010 Sep 8;18(9):1171-83. doi: 10.1016/j.str.2010.06.015. Structure. 2010. PMID: 20826343 - MD simulations and FRET reveal an environment-sensitive conformational plasticity of importin-β.
Halder K, Dölker N, Van Q, Gregor I, Dickmanns A, Baade I, Kehlenbach RH, Ficner R, Enderlein J, Grubmüller H, Neumann H. Halder K, et al. Biophys J. 2015 Jul 21;109(2):277-86. doi: 10.1016/j.bpj.2015.06.014. Biophys J. 2015. PMID: 26200863 Free PMC article. - Impact of the crystallization condition on importin-β conformation.
Tauchert MJ, Hémonnot C, Neumann P, Köster S, Ficner R, Dickmanns A. Tauchert MJ, et al. Acta Crystallogr D Struct Biol. 2016 Jun;72(Pt 6):705-17. doi: 10.1107/S2059798316004940. Epub 2016 May 25. Acta Crystallogr D Struct Biol. 2016. PMID: 27303791 - Mechanics of elastin: molecular mechanism of biological elasticity and its relationship to contraction.
Urry DW, Parker TM. Urry DW, et al. J Muscle Res Cell Motil. 2002;23(5-6):543-59. J Muscle Res Cell Motil. 2002. PMID: 12785104 Review. - Structure, dynamics and interaction with kinase targets: computer simulations of calmodulin.
Yang C, Jas GS, Kuczera K. Yang C, et al. Biochim Biophys Acta. 2004 Mar 11;1697(1-2):289-300. doi: 10.1016/j.bbapap.2003.11.032. Biochim Biophys Acta. 2004. PMID: 15023369 Review.
Cited by
- Mechanisms of intramolecular communication in a hyperthermophilic acylaminoacyl peptidase: a molecular dynamics investigation.
Papaleo E, Renzetti G, Tiberti M. Papaleo E, et al. PLoS One. 2012;7(4):e35686. doi: 10.1371/journal.pone.0035686. Epub 2012 Apr 27. PLoS One. 2012. PMID: 22558199 Free PMC article. - Proteasome activator 200: the heat is on..
Savulescu AF, Glickman MH. Savulescu AF, et al. Mol Cell Proteomics. 2011 May;10(5):R110.006890. doi: 10.1074/mcp.R110.006890. Epub 2011 Mar 9. Mol Cell Proteomics. 2011. PMID: 21389348 Free PMC article. Review. - X-ray crystal structure of the UCS domain-containing UNC-45 myosin chaperone from Drosophila melanogaster.
Lee CF, Hauenstein AV, Fleming JK, Gasper WC, Engelke V, Sankaran B, Bernstein SI, Huxford T. Lee CF, et al. Structure. 2011 Mar 9;19(3):397-408. doi: 10.1016/j.str.2011.01.002. Structure. 2011. PMID: 21397190 Free PMC article. - Disease related single point mutations alter the global dynamics of a tetratricopeptide (TPR) α-solenoid domain.
Llabrés S, Tsenkov MI, MacGowan SA, Barton GJ, Zachariae U. Llabrés S, et al. J Struct Biol. 2020 Jan 1;209(1):107405. doi: 10.1016/j.jsb.2019.107405. Epub 2019 Oct 16. J Struct Biol. 2020. PMID: 31628985 Free PMC article. - Allosteric control of the exportin CRM1 unraveled by crystal structure analysis.
Monecke T, Dickmanns A, Ficner R. Monecke T, et al. FEBS J. 2014 Sep;281(18):4179-94. doi: 10.1111/febs.12842. Epub 2014 Jun 6. FEBS J. 2014. PMID: 24823279 Free PMC article. Review.
References
- Kajander T., Cortajarena A.L., Regan L. A new folding paradigm for repeat proteins. J. Am. Chem. Soc. 2005;127:10188–10190. - PubMed
- Stewart M. Molecular mechanism of the nuclear protein import cycle. Nat. Rev. Mol. Cell Biol. 2007;8:195–208. - PubMed
- Fotin A., Cheng Y., Walz T. Molecular model for a complete clathrin lattice from electron cryomicroscopy. Nature. 2004;432:573–579. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous