Phosphorylation of MLL by ATR is required for execution of mammalian S-phase checkpoint - PubMed (original) (raw)
Phosphorylation of MLL by ATR is required for execution of mammalian S-phase checkpoint
Han Liu et al. Nature. 2010.
Abstract
Cell cycle checkpoints are implemented to safeguard the genome, avoiding the accumulation of genetic errors. Checkpoint loss results in genomic instability and contributes to the evolution of cancer. Among G1-, S-, G2- and M-phase checkpoints, genetic studies indicate the role of an intact S-phase checkpoint in maintaining genome integrity. Although the basic framework of the S-phase checkpoint in multicellular organisms has been outlined, the mechanistic details remain to be elucidated. Human chromosome-11 band-q23 translocations disrupting the MLL gene lead to poor prognostic leukaemias. Here we assign MLL as a novel effector in the mammalian S-phase checkpoint network and identify checkpoint dysfunction as an underlying mechanism of MLL leukaemias. MLL is phosphorylated at serine 516 by ATR in response to genotoxic stress in the S phase, which disrupts its interaction with, and hence its degradation by, the SCF(Skp2) E3 ligase, leading to its accumulation. Stabilized MLL protein accumulates on chromatin, methylates histone H3 lysine 4 at late replication origins and inhibits the loading of CDC45 to delay DNA replication. Cells deficient in MLL showed radioresistant DNA synthesis and chromatid-type genomic abnormalities, indicative of S-phase checkpoint dysfunction. Reconstitution of Mll(-/-) (Mll also known as Mll1) mouse embryonic fibroblasts with wild-type but not S516A or ΔSET mutant MLL rescues the S-phase checkpoint defects. Moreover, murine myeloid progenitor cells carrying an Mll-CBP knock-in allele that mimics human t(11;16) leukaemia show a severe radioresistant DNA synthesis phenotype. MLL fusions function as dominant negative mutants that abrogate the ATR-mediated phosphorylation/stabilization of wild-type MLL on damage to DNA, and thus compromise the S-phase checkpoint. Together, our results identify MLL as a key constituent of the mammalian DNA damage response pathway and show that deregulation of the S-phase checkpoint incurred by MLL translocations probably contributes to the pathogenesis of human MLL leukaemias.
Figures
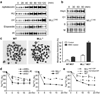
Figure 1. MLL accumulates in S phase upon DNA insults and MLL dysfunction results in S phase checkpoint defects
a, 293T cells were treated with aphidocolin (10 µM), hydroxyurea (1 mM), UV (20 J/m2), etoposide (25 µM), or γ-IR (5 Gy) and then analyzed by anti-MLL immunoblots. b, Synchronized 293T cells were subjected to γ-IR. c, Metaphase spread of MEFs after mitomycin C treatment. Arrowheads indicate chromosomal errors. d, Control- and MLL- knockdown 293T cells (left panel) and wild-type and _MLL_−/− MEFs (right panel) were subjected to RDS assays. e, MPCs were subjected to RDS assays. Data shown in c–e are mean ± s.d. of three independent experiments.
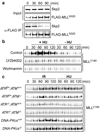
Figure 2. ATR signaling prevents the SCFSkp2-mediated degradation of MLL
a, 293T cells were transfected with the FLAG-MLL expressing construct for 2 days, treated with 1 mM HU for the indicated times, and then subjected to anti-FLAG co-immunoprecipitation assays. Immunoprecipitated FLAG-MLL and co-immunoprecipitated Skp2 were determined by anti-FLAG and anti-Skp2 immunoblots, respectively. b, 293T cells synchronized at mid-S phase were treated with HU in the presence of LY294002 (50 µM) or Wortmannin (10 µM) and then subjected to anti-MLLC180 immunoblots. c, Genetic deletion of ATR prevented the DNA damage-induced accumulation of MLL. MEFs were treated with γ-IR (5 Gy) or HU and then subjected to anti-MLL immunoblots.
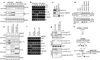
Figure 3. Phosphorylation of MLL at serine 516 by ATR disrupts its interaction with Skp2 and is required for the integrity of S phase checkpoint
a, A diagram of MLL domains. b, 293T cells were transfected with FLAG-MLL for 2 days, treated with 1 mM HU for 1 hour, and then subjected to anti-FLAG co-immunoprecipitation assays. Results were analyzed by immunoblots. c, Recombinant MLL (aa 466–565) was incubated with FLAG-ATR ± FLAG-TopBP1 for in vitro kinase reactions. Results were assessed by anti-phospho-ATM/ATR substrate and anti-phospho-MLL(S516) Western blots. d, FRT+;_MLL_−/− MEFs (Supplementary Fig. 5) reconstituted with wild-type, S516A or ΔSET human MLL were subjected to RDS assays. e, MEFs described in d were subjected to metaphase spread analysis. Data shown in d and e are mean ± s.d. of three independent experiments.
Figure 4. Upon DNA damage MLL accumulates on chromatin to methylate H3K4, resulting in diminished CDC45 loading
a, 293T cells were synchronized in S phase, treated with 25 µM etoposide, fractionated, and subjected to immunoblots using the indicated antibodies. TCE, total cell extracts. b, ChIP assays were performed using the indicated antibodies after etoposide treatment. Immunoprecipitated β-globin replication origin was amplified by PCR. c. Anti-CDC45 immunoprecipitates from 293T nuclear extracts were assessed by anti-histone H3 and anti-H3K4me3 immunoblots. d, The indicated biotin-conjugated histone H3 tails (aa1–21, 2µg) were incubated with nuclear extracts or recombinant GST-CDC45, and then subjected to pull down assays using strepavidin beads. The precipitated H3 peptides were visualized by Coomassie blue stain, whereas co-precipitated CDC45 was detected by anti-CDC45 immunoblots. e, The indicated Jurkat cells were treated with 25 µM etoposide for 90 minutes, fractionated, and then subjected to immunoblots using the indicated antibodies. Of note, MLLC180 denotes endogenous wild-type MLL. f, ChIP assays were performed using the indicated antibodies on Jurkat cells after etoposide treatment. Immunoprecipitated β-globin replication origin was amplified by PCR. g, 293T cells transfected with FLAG-MLL ± FLAG-MLL-AF9 were treated with HU for 90 minutes and then subjected to co-immunoprecipitation assays using the indicated antibodies. h, Model depicts how MLL and MLL-fusions affect the S checkpoint response.
Similar articles
- Role of Dot1-dependent histone H3 methylation in G1 and S phase DNA damage checkpoint functions of Rad9.
Wysocki R, Javaheri A, Allard S, Sha F, Côté J, Kron SJ. Wysocki R, et al. Mol Cell Biol. 2005 Oct;25(19):8430-43. doi: 10.1128/MCB.25.19.8430-8443.2005. Mol Cell Biol. 2005. PMID: 16166626 Free PMC article. - Methylation of histone H4 lysine 20 by PR-Set7 ensures the integrity of late replicating sequence domains in Drosophila.
Li Y, Armstrong RL, Duronio RJ, MacAlpine DM. Li Y, et al. Nucleic Acids Res. 2016 Sep 6;44(15):7204-18. doi: 10.1093/nar/gkw333. Epub 2016 Apr 29. Nucleic Acids Res. 2016. PMID: 27131378 Free PMC article. - Bimodal degradation of MLL by SCFSkp2 and APCCdc20 assures cell cycle execution: a critical regulatory circuit lost in leukemogenic MLL fusions.
Liu H, Cheng EH, Hsieh JJ. Liu H, et al. Genes Dev. 2007 Oct 1;21(19):2385-98. doi: 10.1101/gad.1574507. Genes Dev. 2007. PMID: 17908926 Free PMC article. - MLL translocations, histone modifications and leukaemia stem-cell development.
Krivtsov AV, Armstrong SA. Krivtsov AV, et al. Nat Rev Cancer. 2007 Nov;7(11):823-33. doi: 10.1038/nrc2253. Nat Rev Cancer. 2007. PMID: 17957188 Review. - Interplay between wrn and the checkpoint in s-phase.
Pichierri P. Pichierri P. Ital J Biochem. 2007 Jun;56(2):130-40. Ital J Biochem. 2007. PMID: 17722654 Review.
Cited by
- Targeting the kinase activities of ATR and ATM exhibits antitumoral activity in mouse models of MLL-rearranged AML.
Morgado-Palacin I, Day A, Murga M, Lafarga V, Anton ME, Tubbs A, Chen HT, Ergan A, Anderson R, Bhandoola A, Pike KG, Barlaam B, Cadogan E, Wang X, Pierce AJ, Hubbard C, Armstrong SA, Nussenzweig A, Fernandez-Capetillo O. Morgado-Palacin I, et al. Sci Signal. 2016 Sep 13;9(445):ra91. doi: 10.1126/scisignal.aad8243. Sci Signal. 2016. PMID: 27625305 Free PMC article. - Deubiquitinating enzyme inhibitor alleviates cyclin A1-mediated proteasome inhibitor tolerance in mixed-lineage leukemia.
Ge M, Xu Q, Kang T, Li D, Wang R, Chen Z, Xie S, Wang W, Liu H. Ge M, et al. Cancer Sci. 2021 Jun;112(6):2287-2298. doi: 10.1111/cas.14892. Epub 2021 Apr 5. Cancer Sci. 2021. PMID: 33738896 Free PMC article. - Bromodomain proteins: protectors against endogenous DNA damage and facilitators of genome integrity.
Lee SY, Kim JJ, Miller KM. Lee SY, et al. Exp Mol Med. 2021 Sep;53(9):1268-1277. doi: 10.1038/s12276-021-00673-0. Epub 2021 Sep 21. Exp Mol Med. 2021. PMID: 34548613 Free PMC article. Review. - The Adaptive Mechanisms and Checkpoint Responses to a Stressed DNA Replication Fork.
Saldanha J, Rageul J, Patel JA, Kim H. Saldanha J, et al. Int J Mol Sci. 2023 Jun 22;24(13):10488. doi: 10.3390/ijms241310488. Int J Mol Sci. 2023. PMID: 37445667 Free PMC article. Review. - Mechanisms of Oncogene-Induced Replication Stress: Jigsaw Falling into Place.
Kotsantis P, Petermann E, Boulton SJ. Kotsantis P, et al. Cancer Discov. 2018 May;8(5):537-555. doi: 10.1158/2159-8290.CD-17-1461. Epub 2018 Apr 13. Cancer Discov. 2018. PMID: 29653955 Free PMC article. Review.
References
- Zhou BB, Elledge SJ. The DNA damage response: putting checkpoints in perspective. Nature. 2000;408:433–439. - PubMed
- Kastan MB, Bartek J. Cell-cycle checkpoints and cancer. Nature. 2004;432:316–323. - PubMed
- Bartek J, Lukas C, Lukas J. Checking on DNA damage in S phase. Nat Rev Mol Cell Biol. 2004;5:792–804. - PubMed
- Kolodner RD, Putnam CD, Myung K. Maintenance of genome stability in Saccharomyces cerevisiae. Science. 2002;297:552–557. - PubMed
- Krivtsov AV, Armstrong SA. MLL translocations, histone modifications and leukaemia stem-cell development. Nature reviews. 2007;7:823–833. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 CA123232/CA/NCI NIH HHS/United States
- CA129537/CA/NCI NIH HHS/United States
- R01 CA119008-02/CA/NCI NIH HHS/United States
- R01 CA129537/CA/NCI NIH HHS/United States
- R01 CA119008-03/CA/NCI NIH HHS/United States
- R01 CA119008-05/CA/NCI NIH HHS/United States
- R01 CA119008-04/CA/NCI NIH HHS/United States
- R01 CA119008/CA/NCI NIH HHS/United States
- CA119008/CA/NCI NIH HHS/United States
- CA123232/CA/NCI NIH HHS/United States
- R01 CA119008-01/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous