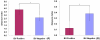Molecular analysis of the diversity of vaginal microbiota associated with bacterial vaginosis - PubMed (original) (raw)
Molecular analysis of the diversity of vaginal microbiota associated with bacterial vaginosis
Zongxin Ling et al. BMC Genomics. 2010.
Abstract
Background: Bacterial vaginosis (BV) is an ecological disorder of the vaginal microbiota that affects millions of women annually, and is associated with numerous adverse health outcomes including pre-term birth and the acquisition of sexually transmitted infections. However, little is known about the overall structure and composition of vaginal microbial communities; most of the earlier studies focused on predominant vaginal bacteria in the process of BV. In the present study, the diversity and richness of vaginal microbiota in 50 BV positive and 50 healthy women from China were investigated using culture-independent PCR-denaturing gradient gel electrophoresis (DGGE) and barcoded 454 pyrosequencing methods, and validated by quantitative PCR.
Results: Our data demonstrated that there was a profound shift in the absolute and relative abundances of bacterial species present in the vagina when comparing populations associated with healthy and diseased conditions. In spite of significant interpersonal variations, the diversity of vaginal microbiota in the two groups could be clearly divided into two clusters. A total of 246,359 high quality pyrosequencing reads was obtained for evaluating bacterial diversity and 24,298 unique sequences represented all phylotypes. The most predominant phyla of bacteria identified in the vagina belonged to Firmicutes, Bacteroidetes, Actinobacteria and Fusobacteria. The higher number of phylotypes in BV positive women over healthy is consistent with the results of previous studies and a large number of low-abundance taxa which were missed in previous studies were revealed. Although no single bacterium could be identified as a specific marker for healthy over diseased conditions, three phyla - Bacteroidetes, Actinobacteria and Fusobacteria, and eight genera including Gardnerella, Atopobium, Megasphaera, Eggerthella, Aerococcus, Leptotrichia/Sneathia, Prevotella and Papillibacter were strongly associated with BV (p < 0.05). These genera are potentially excellent markers and could be used as targets for clinical BV diagnosis by molecular approaches.
Conclusions: The data presented here have clearly profiled the overall structure of vaginal communities and clearly demonstrated that BV is associated with a dramatic increase in the taxonomic richness and diversity of vaginal microbiota. The study also provides the most comprehensive picture of the vaginal community structure and the bacterial ecosystem, and significantly contributes to the current understanding of the etiology of BV.
Figures
Figure 1
Figure 1 PCR-DGGE analysis of the predominant bacterial communities in vaginal swabs from bacterial vaginosis (BV group) and healthy women (CN group). (A) Each lane of the PCR-DGGE gel represented one subject which was selected in its group at random. M represents a marker constructed in this study with the identified bands to facilitate the interpretation of the figure. Bands: 1: Uncultured Sneathia sp.; 2: Fusobacterium nucleatum subsp. nucleatum ATCC 25586; 3: Clostridium thermocellum ATCC 27405; 4: Lactobacillus iners; 5: Clostridium acetobutylicum; 6: Lactobacillus iners; 7: Clostridium thermocellum ATCC 27405; 8: Atopobium vaginae; 9: uncultured Eggerthella sp.; 10: uncultured Megasphaera sp.; 11: Lactobacillus crispatus. (B) Dendrogram of the DGGE profiles shown in panel A.
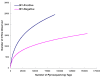
Figure 2
Rarefaction curves were used to estimate richness (in this case the number of taxa at a 3% dissimilarity level) among BV-positive and BV-negative groups. The vertical axis shows the number of OTUs that would be expected to be found after sampling the number of tags or sequences shown on the horizontal axis.
Figure 3
Shannon index and Simpson index were used to estimate diversity (i.e., a combined assessment of the number of 1% dissimilar bacterial taxa and their abundance) among the eight groups. Data shown as mean with SEM. There were significant differences between BV-positive and BV-negative groups by parametric ANOVA.
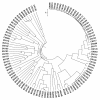
Figure 4
Differentiation in vaginal bacterial communities from 100 individual samples of BV-positive and BV-negative groups (interpersonal variations). Community differentiation was measured by using the unweighted UniFrac algorithm; the scale bar indicates the distance between clusters in UniFrac units. All of the branch nodes shown here were found to be significant (_p <_0.001), indicating that BV-positive and BV-negative harbored distinct bacterial communities.
Figure 5
The relative abundance of vaginal bacterial V3 tags obtained by pyrosequencing from BV-positive and BV-negative individuals, by phylum. Phylogenetic classification for the pyrosequencing analysis obtained from Ribosomal Database Project Classifier analyses. The phyla of Firmicutes, Bacteroidetes, Actinobacteria, Fusobacteria were significant differences between BV-positive and BV-negative groups by parametric ANOVA (_p <_0.000).
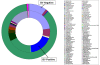
Figure 6
The relative abundance of vaginal bacterial V3 tags obtained by pyrosequencing from BV-positive and BV-negative individuals, by genus and profiled the overall structure of vaginal communities.
Figure 7
Predominant genera detected in vagina from BV-positive and BV-negative individuals. Among these predominant genera, Lactobacillus, Gardnerella, Atopobium, Alloiococcus, Sneathia, Prevotella, Papillibacter, Megasphaera, Eggerthella, Aerococcus were associated with BV significantly (* p < 0.05, ** p < 0.01). Except for Lactobacillus and Alloiococcus, which were detected at a higher level, other genera were more abundant in BV-positive individuals.
Figure 8
Venn diagrams for overlap between BV-positive observed OTUs versus BV-negative observed OTUs. The Venn diagrams show the overlap in all OTUs calculated at the 3% dissimilarity level. The number of species in group BV-positive is 2,455. The number of species in group BV-negative is 1,584. The number of species shared between groups BV-positive and BV-negative is 930. Percentage of species that are shared in groups BV-positive and BV-negative is 29.91%.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases