Elevated O-GlcNAc-dependent signaling through inducible mOGT expression selectively triggers apoptosis - PubMed (original) (raw)
Elevated O-GlcNAc-dependent signaling through inducible mOGT expression selectively triggers apoptosis
Sang-Hoon Shin et al. Amino Acids. 2011 Mar.
Abstract
O-linked N-acetylglucosamine transferase (OGT) catalyzes O-GlcNAc addition to numerous cellular proteins including transcription and nuclear pore complexes and plays a key role in cellular signaling. One differentially spliced isoform of OGT is normally targeted to mitochondria (mOGT) but is quite cytotoxic when expressed in cells compared with the ncOGT isoform. To understand the basis of this selective cytotoxicity, we constructed a fully functional ecdysone-inducible GFP-OGT. Elevated GFP-OGT expression induced a dramatic increase in intracellular O-GlcNAcylated proteins. Furthermore, enhanced OGT expression efficiently triggered programmed cell death. Apoptosis was dependent upon the unique N-terminus of mOGT, and its catalytic activity. Induction of mOGT expression triggered programmed cell death in every cell type tested including INS-1, an insulin-secreting cell line. These studies suggest that deregulated activity of the mitochondrially targeted mOGT may play a role in triggering the programmed cell death observed with diseases such as diabetes mellitus and neurodegeneration.
Figures
Fig. 1
Inducible expression of mOGT in IRES bicistronic vectors leads to toxicity requiring proper targeting and catalytic activity. 1. pI-mOGT-IRES-GFP has the full length OGT gene placed after an ecdysone-inducible promoter and an internal ribosomal entry site is located between OGT and GFP. This vector is used as a backbone for the derivatives described below. 2. pI-mOGT-G-IRES-GFP contains a C-terminal deletion represented by dotted lines (ΔC-term-del). This catalytically defective mOGT replaces the full length wild-type mOGT. 3. pI-mOGT-4A-IRES-GFP contains a deletion (dotted lines) of the 15 N-terminal amino acids thought to be involved in mitochondrial targeting (MLQGHFWLREGIMIS) and has an added histidine tag (solid red stripe) at the N-terminus (ΔN-term-del + His tag). 4. pI-mOGT-4A-G-IRES-GFP is catalytically defective and contains both C-terminal and N-terminal deletions plus the histidine tag described for vectors 2 and 3, respectively. These plasmids were transfected into EcR-293 cells and the surviving colonies were selected with G418 as described. Colonies were counted to quantify the degree of toxicity for each construct. The minus sign means no colonies were observed. There is roughly an order of magnitude difference between one plus and two plus signs. Approximate colony numbers are shown. The colony count was carried out in triplicate and a summary of the three experiments is presented
Fig. 2
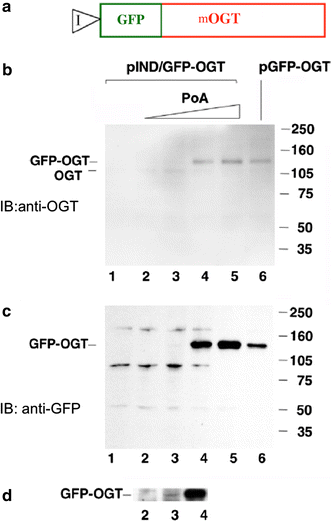
Inducible expression of mOGT in EcR293 cells. a Diagram of the GFP–mOGT construct under the control of an inducible promoter (pIND/GFP–OGT). b Western blot analysis using an anti-OGT antibody to compare expression levels of pIND/GFP–OGT with increasing amounts of Ponasterone A (PoA) to a CMV-driven GFP–OGT fusion (pGFP–OGT). c Western blot analysis of same samples using an anti-GFP antibody. For b and c EcR-293 cells were transfected with pIND/GFP–OGT and incubated without Ponasterone A(PoA) (lane 1), or with the following concentration of PoA: 0.1 µM (lane 2), 0.5 µM (lane 3), 3.0 µM (lane 4), 15.0 µM (lane 5). Cells were also transfected with the CMV-driven plasmid pGFP–OGT (lane 6). On the right side are molecular weight markers. d Longer exposure of the Western blot shown in c between lanes 2 and 4 at 140 kDa range
Fig. 3
_O_-GlcNAcylation is consistent with GFP-fused OGT expression, but not with mutant, catalytically defective, OGT. a Western blot analysis reveals that induction of GFP-fused OGT expression with increasing concentrations of Ponasterone A (PoA) (0.5, 3, 15 µM, respectively) causes an increase in the levels of total _O_-GlcNAc modification. The level of _O_-GlcNAc modified proteins prior to induction is shown in the last lane (−). The anti-_O_-GlcNAc antibody, RL2, was used in the Western Blot analysis. b Immunofluorescent detection of GFP-fused OGT expression and its catalytic activity using RL2 staining. RL2 specifically recognizes _O_-GlcNAcylated proteins. pGFP–OGT-G is the c-terminal deleted, catalytically defective OGT under CMV-driven promoter
Fig. 4
Overexpression of catalytically active GFP–OGT induces apoptosis. a CMV-driven expression of the mitochondrial form of OGT (pGFP–OGT) induces apoptosis as shown by the TUNEL assay. b A catalytically inactive form of OGT (pGFP–OGT-G) does not induce apoptosis. EcR-293 cells were used for transfections in a and b. c and d An insulinoma cell line, INS-1, was transfected with active OGT (pGFP–OGT). The TUNEL assay was used to reveal apoptotic cells. Representative focal planes near the bottom (c) and top (d) of INS-1 cell clusters are shown. Note the apoptotic cells on the outer layer of cells in the clusters. Black and red arrows denote cells scored as positive by the TUNEL assay. e–g EcR-293 cells were transfected with GFP–OGT (pIND/GFP–OGT) and induced with Ponasterone A (PoA) at indicated concentrations. Note the increasing amount of TUNEL-positive cells (black arrows) in g and h compared with the two negative controls e and h pIND/GFP–OGT, no PoA and no plasmid, 15 µM PoA, respectively. i–l Annexin V staining was used to identify apoptotic cells. i EcR-293 cells were treated with 3.0 μM of Ponasterone A to induce expression of pIND/GFP–OGT (green). j Same field stained with an anti-annexin V antibody to reveal apoptotic cells (red). Note near-complete colocalization of GFP–OGT expression and Annexin V staining (compare i and j). k Untreated cells did not stain positive for Annexin V. l As a positive control for apoptosis, 4 μ/ml camptothecin was added to uninduced cells. m DNA ladder assay. Genomic DNA from pIND/GFP–OGT transfected cells (lanes 1–5) was separated on an agarose gel to reveal DNA laddering. Uninduced cells are shown in lane 1. Increasing amounts of Ponasterone A were added at the following concentrations: 0.1 µM (lane 2), 0.5 µM (lane 3), 3.0 µM (lane 4), 15.0 (lane 5). The DNA laddering from CMV-driven expression of pGFP–OGT (lane 6) is comparable to the commercially available positive control from Calf thymus (lane 7). DNA size markers of 100 bp (lane 8) and 1 kb DNA (lane 9) are shown
Similar articles
- Mitochondrial _O_-GlcNAc Transferase (mOGT) Regulates Mitochondrial Structure, Function, and Survival in HeLa Cells.
Sacoman JL, Dagda RY, Burnham-Marusich AR, Dagda RK, Berninsone PM. Sacoman JL, et al. J Biol Chem. 2017 Mar 17;292(11):4499-4518. doi: 10.1074/jbc.M116.726752. Epub 2017 Jan 18. J Biol Chem. 2017. PMID: 28100784 Free PMC article. - Recombinant O-GlcNAc transferase isoforms: identification of O-GlcNAcase, yes tyrosine kinase, and tau as isoform-specific substrates.
Lazarus BD, Love DC, Hanover JA. Lazarus BD, et al. Glycobiology. 2006 May;16(5):415-21. doi: 10.1093/glycob/cwj078. Epub 2006 Jan 23. Glycobiology. 2006. PMID: 16434389 - The mitochondrial O-linked N-acetylglucosamine transferase (mOGT) in the diabetic patient could be the initial trigger to develop Alzheimer disease.
Lozano L, Lara-Lemus R, Zenteno E, Alvarado-Vásquez N. Lozano L, et al. Exp Gerontol. 2014 Oct;58:198-202. doi: 10.1016/j.exger.2014.08.008. Epub 2014 Aug 19. Exp Gerontol. 2014. PMID: 25148700 - O-GlcNAc cycling: implications for neurodegenerative disorders.
Lazarus BD, Love DC, Hanover JA. Lazarus BD, et al. Int J Biochem Cell Biol. 2009 Nov;41(11):2134-46. doi: 10.1016/j.biocel.2009.03.008. Epub 2009 Mar 27. Int J Biochem Cell Biol. 2009. PMID: 19782947 Free PMC article. Review. - Overview of the Assays to Probe _O_-Linked β-_N_-Acetylglucosamine Transferase Binding and Activity.
Balsollier C, Pieters RJ, Anderluh M. Balsollier C, et al. Molecules. 2021 Feb 16;26(4):1037. doi: 10.3390/molecules26041037. Molecules. 2021. PMID: 33669256 Free PMC article. Review.
Cited by
- A little sugar goes a long way: the cell biology of O-GlcNAc.
Bond MR, Hanover JA. Bond MR, et al. J Cell Biol. 2015 Mar 30;208(7):869-80. doi: 10.1083/jcb.201501101. J Cell Biol. 2015. PMID: 25825515 Free PMC article. Review. - Cracking the O-GlcNAc code in metabolism.
Ruan HB, Singh JP, Li MD, Wu J, Yang X. Ruan HB, et al. Trends Endocrinol Metab. 2013 Jun;24(6):301-9. doi: 10.1016/j.tem.2013.02.002. Epub 2013 May 4. Trends Endocrinol Metab. 2013. PMID: 23647930 Free PMC article. Review. - Post-translational modifications in mitochondria: protein signaling in the powerhouse.
Stram AR, Payne RM. Stram AR, et al. Cell Mol Life Sci. 2016 Nov;73(21):4063-73. doi: 10.1007/s00018-016-2280-4. Epub 2016 May 27. Cell Mol Life Sci. 2016. PMID: 27233499 Free PMC article. Review. - Bittersweet memories: linking metabolism to epigenetics through O-GlcNAcylation.
Hanover JA, Krause MW, Love DC. Hanover JA, et al. Nat Rev Mol Cell Biol. 2012 Apr 23;13(5):312-21. doi: 10.1038/nrm3334. Nat Rev Mol Cell Biol. 2012. PMID: 22522719 Review. - Altering O-linked β-N-acetylglucosamine cycling disrupts mitochondrial function.
Tan EP, Villar MT, E L, Lu J, Selfridge JE, Artigues A, Swerdlow RH, Slawson C. Tan EP, et al. J Biol Chem. 2014 May 23;289(21):14719-30. doi: 10.1074/jbc.M113.525790. Epub 2014 Apr 8. J Biol Chem. 2014. PMID: 24713701 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous