Global control of motor neuron topography mediated by the repressive actions of a single hox gene - PubMed (original) (raw)
. 2010 Sep 9;67(5):781-96.
doi: 10.1016/j.neuron.2010.08.008.
Julie Lacombe, Esteban O Mazzoni, Karel F Liem Jr, Jonathan Grinstein, Shaun Mahony, Debnath Mukhopadhyay, David K Gifford, Richard A Young, Kathryn V Anderson, Hynek Wichterle, Jeremy S Dasen
Affiliations
- PMID: 20826310
- PMCID: PMC2955411
- DOI: 10.1016/j.neuron.2010.08.008
Global control of motor neuron topography mediated by the repressive actions of a single hox gene
Heekyung Jung et al. Neuron. 2010.
Abstract
In the developing spinal cord, regional and combinatorial activities of Hox transcription factors are critical in controlling motor neuron fates along the rostrocaudal axis, exemplified by the precise pattern of limb innervation by more than fifty Hox-dependent motor pools. The mechanisms by which motor neuron diversity is constrained to limb levels are, however, not well understood. We show that a single Hox gene, Hoxc9, has an essential role in organizing the motor system through global repressive activities. Hoxc9 is required for the generation of thoracic motor columns, and in its absence, neurons acquire the fates of limb-innervating populations. Unexpectedly, multiple Hox genes are derepressed in Hoxc9 mutants, leading to motor pool disorganization and alterations in the connections by thoracic and forelimb-level subtypes. Genome-wide analysis of Hoxc9 binding suggests that this mode of repression is mediated by direct interactions with Hox regulatory elements, independent of chromatin marks typically associated with repressed Hox genes.
2010 Elsevier Inc. All rights reserved.
Figures
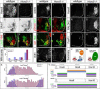
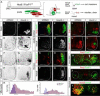
Figure 1. Transformation of Columnar Identities in Hoxc9 Mutants
(A–B) Loss of Hoxc9 protein at thoracic levels in Hoxc9 mutants. Sections show ventral right quadrant of e11.5 spinal cord. (C–F) Expression of VAchT and the number of Lhx3+Hb9+ MMC MNs are grossly normal in Hoxc9 mutants. (G) Quantification of MN columnar subtypes (n>3 mice, error bars represent SEM). In Hoxc9 mutants total MN number at thoracic levels is increased ~30%, approximating limb-level numbers (data not shown). (H–K) Loss of nNOS and pSmad expression in Hoxc9 mutants. (L–O) In the absence of Hoxc9, the number of Isl1/2+ Hb9+ MNs is reduced and Er81 is not detected. (P–U) Ectopic Hoxc6, RALDH2, and FoxP1high MNs at thoracic levels in Hoxc9 mutants. (V–W) Schematic representation of thoracic MN columnar subtypes in wildtype and Hoxc9 mutants. Motor neuron markers for profiling are shown. (X) Quantification of FoxP1+Hoxc6+ and FoxP1+RALDH2+ LMC MNs along the rostrocaudal axis at e11.5. Results show cell counts for one embryo that are typical of n>5 animals. FoxP1 counts represent ventral lateral MNs that express high levels. (Y) Summary of MN columnar transformations in Hoxc9 mutants.
Figure 2. Altered Motor Axon Projection Patterns in Hoxc9 Mutants
(A–D) Vibratome sections showing motor axon projections in wildtype and Hoxc9 mutant embryos at e13.5. (A–B) Axonal projections at brachial levels in wildtype and Hoxc9 mutants. Projections to limb (LMC) and axial muscles (MMC) are preserved. (C–D) In Hoxc9 mutants, axonal projections to sympathetic chain ganglia (scg) are significantly reduced at e13.5 (arrows). See also Figure S3. Vibratome sections show GFP+ motor axons in green, Isl1/2+ scg and dorsal root ganglion (drg) neurons in red. (E–F) Schematic representations of axonal projections of thoracic MNs in wildtype and Hoxc9 mutants. (G–H) The thickness of the intercostal nerves is increased in Hoxc9 mutants (white bars). (I–N) Retrograde labeling of MNs after rhodamine (RhD) injection into intercostal nerves. Ectopic FoxP1high LMC neurons are labeled in Hoxc9 mutants, while Lhx3+ MMC MNs are not labeled.
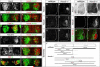
Figure 3. Cell Autonomous Role of Hoxc9 in MN Fate and Hox Gene Expression
(A–L) Analysis of Hoxc9 knockdown at thoracic levels after dsRNA electroporations in chick neural tube. Bolt indicates electroporated side. (A) Hoxc9 dsRNA reduces Hoxc9 protein expression. (B) Nuclear LacZ expression plasmid was coelectroporated to mark electroporated cells. Note that the LacZ plasmid labels only a fraction of cells that incorporate the dsRNA. (C) Hoxc9 dsRNA does not affect Isl1/2 expression. (D) Ectopic RALDH2 is detected after thoracic Hoxc9 RNAi. (E) Loss of pSmad expression. (F) The number of Hb9+Isl1/2+ HMC neurons is reduced after Hoxc9 removal. (G–L) Hoxc6, Hoxa4, Hoxc4, Hoxa5, Hoxa7, and Hoxc8 are ectopically expressed or upregulated in cells that have lost Hoxc9. (M–V) Derepression of Hoxc4, Hoxc5, Hoxa5, Hoxa7, and Hoxc8 expression at thoracic levels in Hoxc9 mutants. The normal brachial patterns of Hox genes were intact in Hoxc9 mutants (Figure S4A–S4H). Ectopic Hox5 expression was relatively weak at thoracic levels, possibly due to the presence of Hoxc8 which normally restricts Hox5 genes to rostral brachial MNs (Dasen et al., 2005). (W–X) Loss of Hoxd9 expression in MNs that ectopically express Hoxc6. (Y) Summary indicating brachially-restricted Hox genes that are ectopically expressed or upregulated in Hoxc9 mutants. Light-gray bars indicated reduced protein expression levels.
Figure 4. Hoxc9 Represses Brachial Hox Genes and LMC Identity
(A–J) Brachial analysis of Hox profiles and motor neuron fates in e12.5 Hb9_∷_Hoxc9 embryos. Hoxc4, Hoxa5, Hoxc6, Hoxc8, and Hoxa7 expression are repressed or significantly downregulated by Hoxc9 in brachial MNs. Hox expression is preserved in the surrounding ventral interneurons. Red dashed line in panel (J) outlines region where Hoxc9 is misexpressed and corresponding region in control mice (I). (K–L) Hb9+ Hoxc9+ MNs are generated in Hb9_∷_Hoxc9 transgenic mice. (M–N) The number of Hb9+Isl1/2+ neurons is increased in Hb9_∷_Hoxc9 transgenic mice. In the absence of a Hox-induced program, MNs appear to remain in a HMC-like ground state. (O–R) Hoxc9 expression in brachial MNs reduces the number of FoxP1+ and RALDH2+ LMC MNs. (S–T) Hoxc9 expression blocks expression of the motor pool marker Pea3.
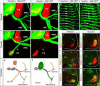
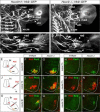
Figure 5. Motor Pool Reorganization in Hoxc9 Mutants
(A) Schematic of the combinatorial Hox codes for motor pools at caudal brachial levels of the spinal cord. Pea3 marks cutaneous maximus (CM) MNs, Scip marks median (med) and ulnar (uln) MNs. Scip MNs are present at the most caudal brachial regions and require exclusion of Hoxc6. (B–I) Multiple markers of the CM pool are detected at thoracic levels in Hoxc9 mutants. The normal brachial patterns were preserved (Figure S6A–S6B and S6I–S6N). (J–M) At caudal brachial levels, upregulation of Hoxc6 expression in Hoxc9 mutants is accompanied by loss of brachial Scip LMC MNs. (N–Q) Altered position of the Scip pool. In Hoxc9 mutants Scip is expressed at thoracic levels. Ectopic Scip neurons are also detected in Hoxc9 RNAi ablated embryos (Figure S6S–S6T). (R) Cell counts for Pea3 and Scip MNs in wildtype and Hoxc9 mutants. Error bars represent n>3 Hoxc9 mutants. (S–T) Ectopic Scip+ and Pea3+ MNs at thoracic levels are clustered normally in Hoxc9 mutants. (U–V) Expression of Meis1/2 is excluded from the Pea3 pool. (W) Pea3 is ectopically expressed at thoracic levels after Hoxc9 RNAi. Bolt: electroporated side. (X) Knockdown of both Hoxc8 + Hoxc9 by dsRNA show that ectopic LMC neurons (FoxP1high) fail to generate ectopic Pea3 at thoracic levels. (Y) Loss of both Hoxc8 and Hoxc9 proteins after coelectroporation of dsRNAs.
Figure 6. Altered Limb Innervation Patterns in Hoxc9 Mutants
(A–D) Forelimb innervation in Hoxc9+/+; Hb9_∷_GFP and _Hoxc9_−/−; Hb9_∷_GFP embryos. Motor axons are visualized by whole mount GFP staining. (A–B) At e12.5 both ulnar (uln) and median (med) nerves show a reduction in length in Hoxc9 mutants. Musculocutaneous (mus), radial (rad), and cutaneous maximus (cm) nerves are similar to wildtype in Hoxc9 mutants. See also Figure S7G–S7J. (C–D) At e13.5 the density of ulnar projections are reduced and there is a loss of the distal branch of the median nerve in Hoxc9 mutants. (E–I) Labeled MNs after RhD injection into the CM nerves. RhD labels the normal Pea3 domain at caudal brachial levels in Hoxc9 mutants. (J–N) Ectopic Pea3 and Scip are labeled after RhD injection into the intercostal nerves at thoracic levels in Hoxc9 mutants. (O–S) In Hoxc9 mutants RhD labels scattered Scip− cells at caudal brachial region, but not ectopic Scip+ cells present at thoracic levels after ulnar injection.
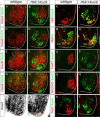
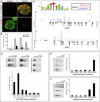
Figure 7. Genomic Analysis of Hoxc9 Binding at Hox Loci in Motor Neurons
(A) Immunostaining showing induction of epitope (V5)-tagged Hoxc9 in embryonic bodies represses Hoxc4 and Hoxa5 expression. (B) RT-PCR analysis of Hoxa4, Hoxa5, Hoxc4, and Hoxc5 transcripts in control and Hoxc9 induced (iHoxc9) ES-cell derived MNs. (C) ChIP-seq signal maps for Hoxc9 binding sites within the Hox-a and Hox-c loci. Hoxc9 consensus motifs are indicated by blue arrowheads and significant binding events are shown in red peak. (D) Hoxc9 binds anterior Hox gene regions at thoracic levels in vivo. Top panels show gel images and bottom panels RT-PCR analysis from ChIP assays. Potential binding sites of Hox genes were assessed by ChIP using Hoxc9 specific antibody. Binding of Hoxc9 to the Hoxa7 3' region was not detected, possibly due to reduced sensitivity of in vivo ChIP assay. Error bars represent standard deviation on triplicates. (E) H3K27me3 chromatin status at Hox gene promoters at thoracic levels (F) H3K27me3 status of Hox gene promoters at brachial levels.
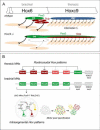
Figure 8. Hox Cross-Repression and Control of Motor Neuron Topography
(A) Summary of alterations in MN organization and muscle innervation in Hoxc9 mutants. HMC MNs are lost and are transformed to an LMC identity. A subset of ectopic thoracic LMC MNs express Pea3 or Scip and project to intercostal muscle. Other motor pool fates may also be acquired by Hox derepression (indicated in gray). As a consequence of the reduction of Scip+ MNs at caudal brachial levels, median and ulnar nerve projections are profoundly reduced. (B) Hoxc9 is a key repressor of brachial Hox genes. In thoracic MNs, Hoxc9 represses brachial Hox genes by directly binding regulatory regions. The efficacy of Hoxc9 repression appears to be graded, where Hox4_–_6 genes are strongly repressed, while Hox7_–_8 gene repression is weaker. Hox10 genes are excluded by the distinct mechanism, likely involving H3K27 methylation-dependent silencing. At brachial levels, an intrasegmental Hox repressor network involving interactions amongst Hox4_–_8 genes determines pool fate on a cell-by-basis (Dasen et al., 2005). At the brachial-thoracic boundary Hoxc6 and Hoxc8 promote LMC fates, defined by high FoxP1 levels and RALDH2. Low levels of Hoxc9 repress Hoxc6 expression to specify the Scip+ LMC pool whereas MNs maintaining both Hoxc6 and Hoxc8 become Pea3+. Pool clustering occurs after MNs have acquired a specific identity.
Similar articles
- Hox repertoires for motor neuron diversity and connectivity gated by a single accessory factor, FoxP1.
Dasen JS, De Camilli A, Wang B, Tucker PW, Jessell TM. Dasen JS, et al. Cell. 2008 Jul 25;134(2):304-16. doi: 10.1016/j.cell.2008.06.019. Cell. 2008. PMID: 18662545 - Genetic and functional modularity of Hox activities in the specification of limb-innervating motor neurons.
Lacombe J, Hanley O, Jung H, Philippidou P, Surmeli G, Grinstein J, Dasen JS. Lacombe J, et al. PLoS Genet. 2013;9(1):e1003184. doi: 10.1371/journal.pgen.1003184. Epub 2013 Jan 24. PLoS Genet. 2013. PMID: 23359544 Free PMC article. - Parallel Pbx-Dependent Pathways Govern the Coalescence and Fate of Motor Columns.
Hanley O, Zewdu R, Cohen LJ, Jung H, Lacombe J, Philippidou P, Lee DH, Selleri L, Dasen JS. Hanley O, et al. Neuron. 2016 Sep 7;91(5):1005-1020. doi: 10.1016/j.neuron.2016.07.043. Epub 2016 Aug 25. Neuron. 2016. PMID: 27568519 Free PMC article. - Transcriptional mechanisms controlling motor neuron diversity and connectivity.
Dalla Torre di Sanguinetto SA, Dasen JS, Arber S. Dalla Torre di Sanguinetto SA, et al. Curr Opin Neurobiol. 2008 Feb;18(1):36-43. doi: 10.1016/j.conb.2008.04.002. Epub 2008 Jun 2. Curr Opin Neurobiol. 2008. PMID: 18524570 Review. - Hox genes and spinal cord development.
Carpenter EM. Carpenter EM. Dev Neurosci. 2002;24(1):24-34. doi: 10.1159/000064943. Dev Neurosci. 2002. PMID: 12145408 Review.
Cited by
- Establishing the Molecular and Functional Diversity of Spinal Motoneurons.
Dasen JS. Dasen JS. Adv Neurobiol. 2022;28:3-44. doi: 10.1007/978-3-031-07167-6_1. Adv Neurobiol. 2022. PMID: 36066819 - Hox transcription factors influence motoneuron identity through the integrated actions of both homeodomain and non-homeodomain regions.
Misra M, Sours E, Lance-Jones C. Misra M, et al. Dev Dyn. 2012 Apr;241(4):718-31. doi: 10.1002/dvdy.23763. Dev Dyn. 2012. PMID: 22411553 Free PMC article. - Dendritic diversification through transcription factor-mediated suppression of alternative morphologies.
Corty MM, Tam J, Grueber WB. Corty MM, et al. Development. 2016 Apr 15;143(8):1351-62. doi: 10.1242/dev.130906. Development. 2016. PMID: 27095495 Free PMC article. - Differential abilities to engage inaccessible chromatin diversify vertebrate Hox binding patterns.
Bulajić M, Srivastava D, Dasen JS, Wichterle H, Mahony S, Mazzoni EO. Bulajić M, et al. Development. 2020 Nov 23;147(22):dev194761. doi: 10.1242/dev.194761. Development. 2020. PMID: 33028607 Free PMC article. - The histone demethylase Kdm6b regulates subtype diversification of mouse spinal motor neurons during development.
Wang W, Cho H, Lee JW, Lee SK. Wang W, et al. Nat Commun. 2022 Feb 17;13(1):958. doi: 10.1038/s41467-022-28636-7. Nat Commun. 2022. PMID: 35177643 Free PMC article.
References
- Alvares LE, Schubert FR, Thorpe C, Mootoosamy RC, Cheng L, Parkyn G, Lumsden A, Dietrich S. Intrinsic, Hox-dependent cues determine the fate of skeletal muscle precursors. Dev Cell. 2003;5:379–390. - PubMed
- Arber S, Han B, Mendelsohn M, Smith M, Jessell TM, Sockanathan S. Requirement for the homeobox gene Hb9 in the consolidation of motor neuron identity. Neuron. 1999;23:659–674. - PubMed
- Ballion B, Morin D, Viala D. Forelimb locomotor generators and quadrupedal locomotion in the neonatal rat. Eur J Neurosci. 2001;14:1727–1738. - PubMed
- Bel-Vialar S, Itasaki N, Krumlauf R. Initiating Hox gene expression: in the early chick neural tube differential sensitivity to FGF and RA signaling subdivides the HoxB genes in two distinct groups. Development. 2002;129:5103–5115. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 NS044385/NS/NINDS NIH HHS/United States
- P01 NS055923/NS/NINDS NIH HHS/United States
- R01 NS062822-01A1/NS/NINDS NIH HHS/United States
- R01 NS062822/NS/NINDS NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous