Determining macromolecular assembly structures by molecular docking and fitting into an electron density map - PubMed (original) (raw)
Determining macromolecular assembly structures by molecular docking and fitting into an electron density map
Keren Lasker et al. Proteins. 2010.
Abstract
Structural models of macromolecular assemblies are instrumental for gaining a mechanistic understanding of cellular processes. Determining these structures is a major challenge for experimental techniques, such as X-ray crystallography, NMR spectroscopy and electron microscopy (EM). Thus, computational modeling techniques, including molecular docking, are required. The development of most molecular docking methods has so far been focused on modeling of binary complexes. We have recently introduced the MultiFit method for modeling the structure of a multisubunit complex by simultaneously optimizing the fit of the model into an EM density map of the entire complex and the shape complementarity between interacting subunits. Here, we report algorithmic advances of the MultiFit method that result in an efficient and accurate assembly of the input subunits into their density map. The successful predictions and the increasing number of complexes being characterized by EM suggests that the CAPRI challenge could be extended to include docking-based modeling of macromolecular assemblies guided by EM.
© 2010 Wiley-Liss, Inc.
Figures
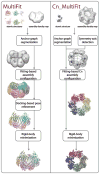
Figure 1. Outline of the MultiFit protocol for simultaneous fitting
The stages of the MultiFit (left) and the Cn_MultiFit (right) algorithms are illustrated from top to bottom. (left) The input is a density map of the MMO hydroxylase complex simulated to 20 Å resolution (gray) and atomic models of the α, β and γ subunits (colors). Segmentation of the density map into 6 regions (light gray) and the corresponding anchor graph (black) as calculated in the “anchor graph segmentation stage”. An assignment of subunits into regions and an atomic model as sampled in the “fitting-based assembly configuration” stage (colors). A refinement of the model (colors) as sampled in the “docking-based pose refinement” stage fitted to the density map (light gray). The final model (colors) superposed on the native complex (gray). (right) The input is an experimentally determined density map of the GroEL complex at 23.5 Å resolution and an atomic structure of the monomeric subunit. The predicted symmetry axis (red) as calculated in the “symmetry axis detection” stage. Segmentation of the density map into 7 regions (light gray) and the corresponding anchor graph (black). A models sampled in the “fitting-based Cn assembly configuration” stage (colors) fitting to the density map (light gray). The final model (colors) superposed on the native complex (gray).
Figure 2. Benchmark results
Final models (colors) for 6 of the benchmark cases. For each test case the PDB entry code, the number of subunits and the final Cα-RMSD to the native structure are listed.
Similar articles
- Building macromolecular assemblies by information-driven docking: introducing the HADDOCK multibody docking server.
Karaca E, Melquiond AS, de Vries SJ, Kastritis PL, Bonvin AM. Karaca E, et al. Mol Cell Proteomics. 2010 Aug;9(8):1784-94. doi: 10.1074/mcp.M000051-MCP201. Epub 2010 Mar 19. Mol Cell Proteomics. 2010. PMID: 20305088 Free PMC article. - MultiFit: a web server for fitting multiple protein structures into their electron microscopy density map.
Tjioe E, Lasker K, Webb B, Wolfson HJ, Sali A. Tjioe E, et al. Nucleic Acids Res. 2011 Jul;39(Web Server issue):W167-70. doi: 10.1093/nar/gkr490. Nucleic Acids Res. 2011. PMID: 21715383 Free PMC article. - Inferential optimization for simultaneous fitting of multiple components into a CryoEM map of their assembly.
Lasker K, Topf M, Sali A, Wolfson HJ. Lasker K, et al. J Mol Biol. 2009 Apr 24;388(1):180-94. doi: 10.1016/j.jmb.2009.02.031. Epub 2009 Feb 20. J Mol Biol. 2009. PMID: 19233204 Free PMC article. - Macromolecular structure comparison and docking: an algorithmic review.
Paquet E, Viktor HL. Paquet E, et al. Curr Pharm Des. 2013;19(12):2183-93. doi: 10.2174/1381612811319120006. Curr Pharm Des. 2013. PMID: 23016846 Review. - Automated Modeling and Validation of Protein Complexes in Cryo-EM Maps.
Cragnolini T, Sweeney A, Topf M. Cragnolini T, et al. Methods Mol Biol. 2021;2215:189-223. doi: 10.1007/978-1-0716-0966-8_9. Methods Mol Biol. 2021. PMID: 33368005 Review.
Cited by
- Mapping the native organization of the yeast nuclear pore complex using nuclear radial intensity measurements.
Vallotton P, Rajoo S, Wojtynek M, Onischenko E, Kralt A, Derrer CP, Weis K. Vallotton P, et al. Proc Natl Acad Sci U S A. 2019 Jul 16;116(29):14606-14613. doi: 10.1073/pnas.1903764116. Epub 2019 Jul 1. Proc Natl Acad Sci U S A. 2019. PMID: 31262825 Free PMC article. - Stoichiometry and compositional plasticity of the yeast nuclear pore complex revealed by quantitative fluorescence microscopy.
Rajoo S, Vallotton P, Onischenko E, Weis K. Rajoo S, et al. Proc Natl Acad Sci U S A. 2018 Apr 24;115(17):E3969-E3977. doi: 10.1073/pnas.1719398115. Epub 2018 Apr 9. Proc Natl Acad Sci U S A. 2018. PMID: 29632211 Free PMC article. - Principles for Integrative Structural Biology Studies.
Rout MP, Sali A. Rout MP, et al. Cell. 2019 May 30;177(6):1384-1403. doi: 10.1016/j.cell.2019.05.016. Cell. 2019. PMID: 31150619 Free PMC article. Review. - UCSF Chimera, MODELLER, and IMP: an integrated modeling system.
Yang Z, Lasker K, Schneidman-Duhovny D, Webb B, Huang CC, Pettersen EF, Goddard TD, Meng EC, Sali A, Ferrin TE. Yang Z, et al. J Struct Biol. 2012 Sep;179(3):269-78. doi: 10.1016/j.jsb.2011.09.006. Epub 2011 Sep 22. J Struct Biol. 2012. PMID: 21963794 Free PMC article. - Computational structure modeling for diverse categories of macromolecular interactions.
Aderinwale T, Christoffer CW, Sarkar D, Alnabati E, Kihara D. Aderinwale T, et al. Curr Opin Struct Biol. 2020 Oct;64:1-8. doi: 10.1016/j.sbi.2020.05.017. Epub 2020 Jun 27. Curr Opin Struct Biol. 2020. PMID: 32599506 Free PMC article. Review.
References
- Alberts B. The cell as a collection of protein machines: preparing the next generation of molecular biologists. Cell. 1998;92(3):291–294. - PubMed
- Robinson CV, Sali A, Baumeister W. Molecular sociology of the cell. Nature. 2007;450:973–982. - PubMed
- Sali A, Glaeser R, Earnest T, Baumeister W. From words to literature in structural proteomics. Nature. 2003;422(6928):216–225. - PubMed
- Alber F, Forster F, Korkin D, Topf M, Sali A. Integrating diverse data for structure determination of macromolecular assemblies. Annu Rev Biochem. 2008;77:443–477. - PubMed
- Pons C, Grosdidier S, Solernou A, Perez-Cano L, Fernandez-Recio J. Present and future challenges and limitations in protein-protein docking. Proteins. 78(1):95–108. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 GM54762/GM/NIGMS NIH HHS/United States
- R01 GM083960/GM/NIGMS NIH HHS/United States
- U54 RR022220/RR/NCRR NIH HHS/United States
- R01 GM083960-04/GM/NIGMS NIH HHS/United States
- R01 GM054762/GM/NIGMS NIH HHS/United States
- PN2 EY016525-05/EY/NEI NIH HHS/United States
- U54 RR022220-05/RR/NCRR NIH HHS/United States
- R01 GM054762-06/GM/NIGMS NIH HHS/United States
- PN2 EY016525/EY/NEI NIH HHS/United States
LinkOut - more resources
Full Text Sources