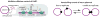Fundamental limits on the suppression of molecular fluctuations - PubMed (original) (raw)
Fundamental limits on the suppression of molecular fluctuations
Ioannis Lestas et al. Nature. 2010.
Abstract
Negative feedback is common in biological processes and can increase a system's stability to internal and external perturbations. But at the molecular level, control loops always involve signalling steps with finite rates for random births and deaths of individual molecules. Here we show, by developing mathematical tools that merge control and information theory with physical chemistry, that seemingly mild constraints on these rates place severe limits on the ability to suppress molecular fluctuations. Specifically, the minimum standard deviation in abundances decreases with the quartic root of the number of signalling events, making it extremely expensive to increase accuracy. Our results are formulated in terms of experimental observables, and existing data show that cells use brute force when noise suppression is essential; for example, regulatory genes are transcribed tens of thousands of times per cell cycle. The theory challenges conventional beliefs about biochemical accuracy and presents an approach to the rigorous analysis of poorly characterized biological systems.
Figures
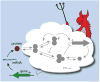
Figure 1. Schematic of optimal control networks and information loss
Biological networks can be overwhelmingly complex, with numerous feedback loops and signaling steps. Predictions about noise then rely on quantitative estimates for how every probabilistic reaction rate responds to every type of perturbation. To investigate bounds on behavior, most of the network is here replaced by a ‘control demon’ representing a controller that is optimized over all possible network topologies, rates and mechanisms. The bounds are then calculated in terms of the few specified features.
Figure 2. Hard limits on standard deviations
(left) Intrinsic noise (Eq. (1)). The lower limit on the relative standard deviation normalized by that of a Poisson distribution, as a function of the ratio _N_2/_N_1. Blue curve corresponds to reaction scheme (1), and red to the autocatalytic scheme described above Eq. (5). The quartic root is the strongest relative response along either curve, while at low relative signaling frequencies the limit is an even more damped function of _N_2/_N_1. (Left, inset) The same lower limit for an average of 100 X1 molecules, as a function of _N_2. (Right) Extrinsic noise. X1 is made at rate x_3_u, where X3 is born with constant probability and decays exponentially with rate 1/_τ_3, while intrinsic birth and death noise in X1 is ignored. For _τ_3≪_τ_1 or _τ_3≫_τ_1, the quartic root asymptotic still applies, essentially because the process mimics a one-variable random process in both cases. At intermediate time-scales the _N_2 dependence is less strict and _τ_3=_τ_1 produces an asymptotic power law exponent of 3/8 rather than ¼, partly supporting previous, conclusions that extrinsic noise is slightly easier to suppress. However, many actual control systems may find intermediately slow noise the hardest to eliminate and any predictions about suppressing extrinsic noise will depend on the properties of that noise. The predicted extrinsic noise limit is also a conservative estimate, and the actual magnitude of the noise limit may be slightly higher (SI).
Figure 3. Plasmid replication control
(Left) Plasmid ColE1 expresses an inhibitor that prevents replication, similarly to the self-replication model in the main text with X1 as plasmid and X2 as inhibitor. Because plasmids are under selection for noise suppression the theory predicts it must maximize expression rates and minimize the length of signaling cascades while still achieving ‘cooperative’ nonlinear effects in the control loop. ColE1 indeed expresses a short-lived anti-sense RNA inhibitor (RNA I) tens of thousands of times per cell cycle (~10Hz), that directly and irreversibly blocks the maturation of a constitutively synthesized sense-RNA replication pre-primer (RNA II) – eliminating both the translation step and binding and unbinding to genes and making it energetically and mechanistically possible to produce inhibitors at such high rates. ColE1 could also create strongly nonlinear control kinetics by exploiting kinetic proofreading in RNA II elongation,. Many unrelated plasmids similarly express anti-sense inhibitors at high rates, avoid cascades, and use multistep inhibition kinetics. (Right) Plasmids such as P1, F, and pSC101 use ‘handcuffing’ mechanisms, where repeated DNA sequences (iterons) bind each other and prevent replication. This can achieve similar homeostatic dynamics as monomer-dimer equilibria where a higher fraction of molecules are in dimer form at higher abundance. Using DNA itself as inhibitor this could eliminate the need for indirect signaling altogether, but because the mechanisms seem incapable of strongly nonlinear corrections, most such plasmids use additional control systems that go through gene expression and thus are subject to information loss. Plasmids also commonly use counteracting loops, where replication inhibitors also auto-inhibit their own synthesis – a counter-intuitive strategy that in fact can improve control greatly (increasing _H_22 for a given high _H_21 in Eq. (4)).
Comment in
- Systems biology: The cost of feedback control.
Sun L, Becskei A. Sun L, et al. Nature. 2010 Sep 9;467(7312):163-4. doi: 10.1038/467163a. Nature. 2010. PMID: 20829785 No abstract available.
Similar articles
- Transient amplification limits noise suppression in biochemical networks.
Dixon J, Lindemann A, McCoy JH. Dixon J, et al. Phys Rev E. 2016 Jan;93(1):012415. doi: 10.1103/PhysRevE.93.012415. Epub 2016 Jan 26. Phys Rev E. 2016. PMID: 26871109 - Macromolecular crowding: chemistry and physics meet biology (Ascona, Switzerland, 10-14 June 2012).
Foffi G, Pastore A, Piazza F, Temussi PA. Foffi G, et al. Phys Biol. 2013 Aug;10(4):040301. doi: 10.1088/1478-3975/10/4/040301. Epub 2013 Aug 2. Phys Biol. 2013. PMID: 23912807 - Interlinked fast and slow positive feedback loops drive reliable cell decisions.
Brandman O, Ferrell JE Jr, Li R, Meyer T. Brandman O, et al. Science. 2005 Oct 21;310(5747):496-8. doi: 10.1126/science.1113834. Science. 2005. PMID: 16239477 Free PMC article. - Physicochemical modelling of cell signalling pathways.
Aldridge BB, Burke JM, Lauffenburger DA, Sorger PK. Aldridge BB, et al. Nat Cell Biol. 2006 Nov;8(11):1195-203. doi: 10.1038/ncb1497. Nat Cell Biol. 2006. PMID: 17060902 Review. - Engineering challenges of BioNEMS: the integration of microfluidics, micro- and nanodevices, models and external control for systems biology.
Wikswo JP, Prokop A, Baudenbacher F, Cliffel D, Csukas B, Velkovsky M. Wikswo JP, et al. IEE Proc Nanobiotechnol. 2006 Aug;153(4):81-101. doi: 10.1049/ip-nbt:20050045. IEE Proc Nanobiotechnol. 2006. PMID: 16948492 Review.
Cited by
- Improved bounds on entropy production in living systems.
Skinner DJ, Dunkel J. Skinner DJ, et al. Proc Natl Acad Sci U S A. 2021 May 4;118(18):e2024300118. doi: 10.1073/pnas.2024300118. Proc Natl Acad Sci U S A. 2021. PMID: 33906948 Free PMC article. - Gene expression noise is affected differentially by feedback in burst frequency and burst size.
Bokes P, Singh A. Bokes P, et al. J Math Biol. 2017 May;74(6):1483-1509. doi: 10.1007/s00285-016-1059-4. Epub 2016 Sep 24. J Math Biol. 2017. PMID: 27665109 - Transcription factor binding kinetics constrain noise suppression via negative feedback.
Grönlund A, Lötstedt P, Elf J. Grönlund A, et al. Nat Commun. 2013;4:1864. doi: 10.1038/ncomms2867. Nat Commun. 2013. PMID: 23673649 - Roles for microRNAs in conferring robustness to biological processes.
Ebert MS, Sharp PA. Ebert MS, et al. Cell. 2012 Apr 27;149(3):515-24. doi: 10.1016/j.cell.2012.04.005. Cell. 2012. PMID: 22541426 Free PMC article. Review. - Increased gene expression noise in human cancers is correlated with low p53 and immune activities as well as late stage cancer.
Han R, Huang G, Wang Y, Xu Y, Hu Y, Jiang W, Wang T, Xiao T, Zheng D. Han R, et al. Oncotarget. 2016 Nov 1;7(44):72011-72020. doi: 10.18632/oncotarget.12457. Oncotarget. 2016. PMID: 27713130 Free PMC article.
References
- Ozbudak EM, Thattai M, Kurtser I, Grossman AD, van Oudenaarden A. Regulation of noise in the expression of a single gene. Nature Genetics. 2002;31:69–73. - PubMed
- Elowitz MB, Levine AJ, Siggia ED, Swain PS. Stochastic gene expression in a single cell. Science (Washington, DC, United States) 2002;297:1183–1186. - PubMed
- Newman JR, et al. Single-cell proteomic analysis of S. cerevisiae reveals the architecture of biological noise. Nature. 2006;441:840–846. - PubMed
- Golding I, Paulsson J, Zawilski SM, Cox EC. Real-time kinetics of gene activity in individual bacteria. Cell (Cambridge, MA, United States) 2005;123:1025–1036. - PubMed
- Paulsson J, Ehrenberg M. Noise in a minimal regulatory network: Plasmid copy number control. Quarterly Reviews of Biophysics. 2001;34:1–59. - PubMed
Publication types
MeSH terms
Grants and funding
- P50 GM068763/GM/NIGMS NIH HHS/United States
- R01 GM081563/GM/NIGMS NIH HHS/United States
- GM068763-06/GM/NIGMS NIH HHS/United States
- BB/C008073/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
LinkOut - more resources
Full Text Sources