RNase L releases a small RNA from HCV RNA that refolds into a potent PAMP - PubMed (original) (raw)
RNase L releases a small RNA from HCV RNA that refolds into a potent PAMP
Krishnamurthy Malathi et al. RNA. 2010 Nov.
Abstract
Triggering and propagating an intracellular innate immune response is essential for control of viral infections. RNase L is a host endoribonuclease and a pivotal component of innate immunity that cleaves viral and cellular RNA within single-stranded loops releasing small structured RNAs with 5'-hydroxyl (5'-OH) and 3'-monophosphoryl (3'-p) groups. In 2007, we reported that RNase L cleaves self RNA to produce small RNAs that function as pathogen-associated molecular patterns (PAMPs). However, the precise sequence and structure of PAMP RNAs produced by RNase L is unknown. Here we used hepatitis C virus RNA as substrate to characterize RNase L mediated cleavage products [named suppressor of virus RNA (svRNA)] for their ability to activate RIG-I like receptors (RLR). The NS5B region of HCV RNA was cleaved by RNase L to release an svRNA that bound to RIG-I, displacing its repressor domain and stimulating its ATPase activity while signaling to the IFN-β gene in intact cells. All three of these RIG-I functions were dependent on the presence in svRNA of the 3'-p. Furthermore, svRNA suppressed HCV replication in vitro through a mechanism involving IFN production and triggered a RIG-I-dependent hepatic innate immune response in mice. RNase L and OAS (required for its activation) were both expressed in hepatocytes from HCV-infected patients, raising the possibility that the OAS/RNase L pathway might suppress HCV replication in vivo. It is proposed that RNase L mediated cleavage of HCV RNA generates svRNA that activates RIG-I, thus propagating innate immune signaling to the IFN-β gene.
Figures
FIGURE 1.
Cleavage of HCV RNA by RNase L produces small RNAs with PAMP activity. (A) Activation of the IFN-β promoter after 18 h in Huh7 cells in response to transfection with undigested or RNase L-digested HCV RNA segments. (B) RNase L releases svRNA3 from two adjacent stem–loops in the NS5B region of the HCV open reading frame. (C) Detection of RNase L-mediated cleavage product of HCV RNA (svRNA3) in Huh7.5 cells at 96 h after electroporated with full-length HCV genomic RNA or a region of HCV RNA encoding svRNA3 (nt 8703–9416) as determined in a Northern blot (upper) (Materials and Methods). Small RNAs, <200 nt (10 μg), and 100 ng of svRNA3 were stained with gelstar (Cambrex Bio Science) (lower).
FIGURE 2.
Activation of RIG-I ATPase activity and IFN-β induction by svRNA3 and its derivatives. Predicted RNA secondary structure of (A) svRNA3 (5′-p3/3′-OH); (B) svRNA3 (5′-OH/3′-p); (C) CsvRNA3 (5′-p3/3′-OH), complement of svRNA3; (D) svRNA3 short (5′-OH/3′-OH or 5′-OH/3′-p), the main stem–loop structure of svRNA3; (E) 5′ΔsvRNA3 (5′-p3/3′-OH), svRNA3 deleted for the 5′-ss overhang; (F) 3′ΔsvRNA3 (5′-p3/3′-OH), svRNA3 deleted for the 3′-ss overhang; (G) 5′ 3′ΔsvRNA3 (5′-p3/3′-OH), svRNA3 deleted for both the 3′- and 5′-ss overhangs. (H) Activation of RIG-I or MDA5 ATPase (20 min at 37°C) by the indicated RNAs. (I) Activation of the IFN-β promoter in Huh7 cells after 18 h by transfection of the indicated RNAs. (J) Induction of IFN-β by svRNA3 in mouse embryo fibroblasts (mef). IFN-β levels in supernatants of WT, RIG-I-deficient, MDA5-deficient, or IPS1-deficient mouse embryo fibroblasts 18 h after transfection with 30 pmol of svRNA3. IFN-β protein levels were measured by ELISA. RNA structures were as predicted by mfold software (Zuker 2003).
FIGURE 3.
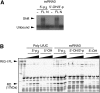
SvRNA3 binds RIG-I causing the release of its repressor domain (RD). (A) RIG-I binding of svRNA3 with different termini (as shown) as determined by gel-shift analysis of full-length (FL) RIG-I or its N-terminal (N) polypeptide (1–228 amino acids). (B) Release of RIG-I RD by partial trypsin digestion. The silver-stained gel image shows trypsin digestion products of Flag-RIG-I incubated with 3, 6, 15, or 30 pmol of the indicated RNA.
FIGURE 4.
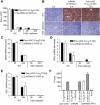
HCV svRNA3 induces a hepatic innate immune response and suppresses HCV replication by a paracrine mechanism. (A–E) Hydrodynamic tail vein injection of poly-U/UC (5′-p3/3′-OH) or svRNA3 (5′-OH/3′-p) in WT and _Rig-I_−/− mice (n = 3). (A) Induction of circulating IFN-β by poly-U/UC or svRNA3. (B) Hepatic induction of ISG54 protein as determined by immunohistochemistry (IHC) by poly-U/UC or svRNA3. (C–E) Hepatic induction of Isg56, Ifnb, and Rig-i mRNAs by poly-U/UC or svRNA3 as determined by qRT-PCR. (F) Paracrine inhibition of HCV infections by poly-U/UC (5′-p3/3′-OH) or svRNA3 (5′-OH/3′-p). Huh7.5 cells (in triplicate) were treated for 12 h before HCV infection with medium containing IFN-β or conditioned medium from Huh7 cells transfected with the indicated RNAs. The graph shows the number of infected cells as determined by focus-forming unit (FFU) assay at 48 h after infection.
FIGURE 5.
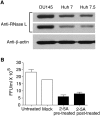
Inhibition of HCV replication in response to 2-5A activation of RNase L. (A) RNase L levels in lysates (30 μg) of DU145 prostate cancer cells and hepatoma cell lines Huh7 and Huh7.5 as determined in immunoblots (different exposures of the same blot are shown). Blots were probed with monoclonal antibody reactive to human RNase L (Dong and Silverman 1995) and compared to anti-β-actin as control. (B) Huh7 cells were either mock transfected or transfected with 2-5A trimer (p35′A2′p5′A2′p5′A) before or after HCV infection (JFH1 strain) for 8 h. Supernatants were harvested at 48 h post-infection and titered for HCV by focus-forming assays.
FIGURE 6.
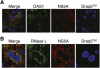
Hepatic expression of HCV NS5A, OAS1, and RNase L in HCV-infected liver. Images show a 0.15 μm optical section of liver biopsy specimen stained with Draq-5 to show nuclei (blue), and immunostained with antibodies specific to NS5A (red) and (A) OAS1 (green) or (B) RNase-L (green). The left panel of each set features the merged image showing sites of protein codistribution (yellow). Images represent a single patient and are representative of analyses from six different patients with chronic HCV infection (data not shown). 60X magnification.
FIGURE 7.
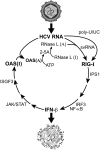
Hypothetical temporal appearance and roles of the PAMPs, poly-U/UC, and svRNA3, in the innate immune response against HCV infections. Active (A) and inactive (I) forms of OAS and RNase L are indicated.
Similar articles
- Uridine composition of the poly-U/UC tract of HCV RNA defines non-self recognition by RIG-I.
Schnell G, Loo YM, Marcotrigiano J, Gale M Jr. Schnell G, et al. PLoS Pathog. 2012;8(8):e1002839. doi: 10.1371/journal.ppat.1002839. Epub 2012 Aug 2. PLoS Pathog. 2012. PMID: 22912574 Free PMC article. - RNase L Amplifies Interferon Signaling by Inducing Protein Kinase R-Mediated Antiviral Stress Granules.
Manivannan P, Siddiqui MA, Malathi K. Manivannan P, et al. J Virol. 2020 Jun 16;94(13):e00205-20. doi: 10.1128/JVI.00205-20. Print 2020 Jun 16. J Virol. 2020. PMID: 32295917 Free PMC article. - Innate immunity induced by composition-dependent RIG-I recognition of hepatitis C virus RNA.
Saito T, Owen DM, Jiang F, Marcotrigiano J, Gale M Jr. Saito T, et al. Nature. 2008 Jul 24;454(7203):523-7. doi: 10.1038/nature07106. Epub 2008 Jun 11. Nature. 2008. PMID: 18548002 Free PMC article. - Links between recognition and degradation of cytoplasmic viral RNA in innate immune response.
Oshiumi H, Mifsud EJ, Daito T. Oshiumi H, et al. Rev Med Virol. 2016 Mar;26(2):90-101. doi: 10.1002/rmv.1865. Epub 2015 Dec 8. Rev Med Virol. 2016. PMID: 26643446 Review. - New insights into the role of RNase L in innate immunity.
Chakrabarti A, Jha BK, Silverman RH. Chakrabarti A, et al. J Interferon Cytokine Res. 2011 Jan;31(1):49-57. doi: 10.1089/jir.2010.0120. Epub 2010 Dec 29. J Interferon Cytokine Res. 2011. PMID: 21190483 Free PMC article. Review.
Cited by
- Hepatocyte Intrinsic Innate Antiviral Immunity against Hepatitis Delta Virus Infection: The Voices of Bona Fide Human Hepatocytes.
Woo Y, Ma M, Okawa M, Saito T. Woo Y, et al. Viruses. 2024 May 8;16(5):740. doi: 10.3390/v16050740. Viruses. 2024. PMID: 38793622 Free PMC article. Review. - Unraveling the Role of RNase L Knockout in Alleviating Immune Response Activation in Mice Bone Marrow after Irradiation.
Ding K, Li H, Tai F, Duan J, Wang Q, Zhai R, Fu H, Ge C, Zheng X. Ding K, et al. Int J Mol Sci. 2024 Feb 27;25(5):2722. doi: 10.3390/ijms25052722. Int J Mol Sci. 2024. PMID: 38473966 Free PMC article. - Orbivirus NS4 Proteins Play Multiple Roles to Dampen Cellular Responses.
Mohd Jaafar F, Belhouchet M, Monsion B, Bell-Sakyi L, Mertens PPC, Attoui H. Mohd Jaafar F, et al. Viruses. 2023 Sep 12;15(9):1908. doi: 10.3390/v15091908. Viruses. 2023. PMID: 37766314 Free PMC article. - Crosstalk between Autophagy and RLR Signaling.
Ke PY. Ke PY. Cells. 2023 Mar 21;12(6):956. doi: 10.3390/cells12060956. Cells. 2023. PMID: 36980296 Free PMC article. Review. - Phenotypic and Transcriptional Changes of Pulmonary Immune Responses in Dogs Following Canine Distemper Virus Infection.
Chludzinski E, Klemens J, Ciurkiewicz M, Geffers R, Pöpperl P, Stoff M, Shin DL, Herrler G, Beineke A. Chludzinski E, et al. Int J Mol Sci. 2022 Sep 2;23(17):10019. doi: 10.3390/ijms231710019. Int J Mol Sci. 2022. PMID: 36077417 Free PMC article.
References
- Armstrong GL, Wasley A, Simard EP, McQuillan GM, Kuhnert WL, Alter MJ 2006. The prevalence of hepatitis C virus infection in the United States, 1999 through 2002. Ann Intern Med 144: 705–714 - PubMed
- Cole JL, Carroll SS, Kuo LC 1996. Stoichiometry of 2′,5′-oligoadenylate-induced dimerization of ribonuclease L. A sedimentation equilibrium study. J Biol Chem 271: 3979–3981 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R15 AI089518/AI/NIAID NIH HHS/United States
- AI060389/AI/NIAID NIH HHS/United States
- DA024563/DA/NIDA NIH HHS/United States
- R01 AI060389/AI/NIAID NIH HHS/United States
- R01 DA024563/DA/NIDA NIH HHS/United States
- CA44059/CA/NCI NIH HHS/United States
- 1RC1A1086041/RC/CCR NIH HHS/United States
- RC1 AI086041/AI/NIAID NIH HHS/United States
- R01 CA044059/CA/NCI NIH HHS/United States
- R56 AI060389/AI/NIAID NIH HHS/United States
- R15 AI089518-01/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous