The psychiatric disease risk factors DISC1 and TNIK interact to regulate synapse composition and function - PubMed (original) (raw)
. 2011 Oct;16(10):1006-23.
doi: 10.1038/mp.2010.87. Epub 2010 Sep 14.
E I Charych, V L Pulito, J B Lee, N M Graziane, R A Crozier, R Revilla-Sanchez, M P Kelly, A J Dunlop, H Murdoch, N Taylor, Y Xie, M Pausch, A Hayashi-Takagi, K Ishizuka, S Seshadri, B Bates, K Kariya, A Sawa, R J Weinberg, S J Moss, M D Houslay, Z Yan, N J Brandon
Affiliations
- PMID: 20838393
- PMCID: PMC3176992
- DOI: 10.1038/mp.2010.87
The psychiatric disease risk factors DISC1 and TNIK interact to regulate synapse composition and function
Q Wang et al. Mol Psychiatry. 2011 Oct.
Abstract
Disrupted in schizophrenia 1 (DISC1), a genetic risk factor for multiple serious psychiatric diseases including schizophrenia, bipolar disorder and autism, is a key regulator of multiple neuronal functions linked to both normal development and disease processes. As these diseases are thought to share a common deficit in synaptic function and architecture, we have analyzed the role of DISC1 using an approach that focuses on understanding the protein-protein interactions of DISC1 specifically at synapses. We identify the Traf2 and Nck-interacting kinase (TNIK), an emerging risk factor itself for disease, as a key synaptic partner for DISC1, and provide evidence that the DISC1-TNIK interaction regulates synaptic composition and activity by stabilizing the levels of key postsynaptic density proteins. Understanding the novel DISC1-TNIK interaction is likely to provide insights into the etiology and underlying synaptic deficits found in major psychiatric diseases.
Conflict of interest statement
Conflict of interest
The authors declare no conflict of interest.
Figures
Figure 1
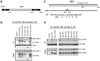
A 13-amino acid region on DISC1 is required for its interaction with TNIK. (a) Schematic representation of TNIK with the N-terminal kinase domain (KD) and C-terminal citron homology domain (CNH). The point mutant K54R is a kinase-dead mutant. (b) DISC1 binds to the kinase domain of TNIK. Myc-DISC1 co-immunoprecipitates with HA-TNIK WT but not HA-TNIK ΔKD from HEK293 cells. (c) Schematic representation of DISC1 and point mutants within the TNIK binding site. Amino acids 329–350 are deleted in DISC1 ΔTNIK. DISC1 AS1–6 are alanine substitution (AS) mutants with the underlined amino acid residues mutated to alanine. (d) Amino acids crucial for the interaction of DISC1 and TNIK. Myc-DISC1 WT, but not myc-DISC1 DTNIK, co-immunoprecipitates with HA-TNIK from HEK293 cells.
Figure 2
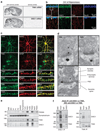
DISC1 associates with TNIK in the brain. (a) Expression of TNIK and DISC1 mRNA in adult mouse brain shown by in situ hybridization. (b) Single plane confocal images of CA1 from adult mice immunostained for DISC1 (green), TNIK (red) and the neuronal marker NeuN (blue) showing colocalization of DISC1 and TNIK in likely dendritic projections. (c) Single plane confocal images of 19–26 DIV rat primary hippocampal cultures showing colocalization of TNIK with neuronal and PSD markers and DISC1, but not with the glial marker, glial fibrillary acidic protein (GFAP). (d) Electron micrographs, with pre-embedding immunogold staining, showing the presence of TNIK and DISC1 in adult rat CA1 dendritic spines. (e) TNIK and DISC1 are enriched in PSD fractions from rat hippocampi. BH, brain homogenate; ERGO, ER and Golgi apparatus; P1, nuclei and unbroken cells; P2, organelles; PSD1–3, PSD fractions 1–3; S1, cytoplasm; SarS, Sarcosyl soluble fraction; TxS1 and 2, Triton X-100 soluble fractions 1 and 2. (f) Co-immunoprecipitation of TNIK and DISC1 from rat brain. IPs were performed with DISC1-440 (left) and TNIK Santa Cruz (Right) antibodies, followed by western blotting (WB) using both antibodies.
Figure 3
DISC1 inhibits TNIK kinase activity-dependent cellular outputs. (a) Kinase-dependent cell rounding induced by TNIK. NIH3T3 cells were transfected with HA-TNIK WT, K54R or Myc-DISC1 WT alone, followed by immunofluorescence staining with anti-HA or anti-Myc antibody. (b) DISC1 inhibits TNIK-induced cell rounding. NIH3T3 cells were co-transfected with HA-TNIK WT and Myc-DISC1 WT followed by immunofluorescence staining with anti-HA and anti-Myc antibodies. (c) The TNIK binding site on DISC1 is required for inhibition of TNIK-induced cell rounding. Representative images of cells with indicated co-transfection and stained for HA-TNIK, and the length-to-breadth ration of cells were analyzed in (d) as a surrogate measure of cell spreading. _N_>30. *P<0.05, **P<0.01, ***P<0.001.
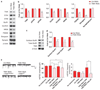
Figure 4
The binding site-derived DISC1 peptide inhibits TNIK kinase activity. (a) Sequences of DISC1 peptides fused with the membrane transduction domain of Tat. DISCpep-WT contains amino acids 329–349 of human DISC1 and DISCpep-PM has three residues substituted by alanine. Mouse/rat version of DISCpep-WT contains amino acids 330–350 in mouse DISC1 and is 100% identical in mouse and rat. (b) DISCpep-WT inhibits kinase activity of TNIK in vitro. Kinase reactions were carried out using MBP as substrate with 0.15 µM of purified human TNIK and 10 µM peptides with or without a TNIK pre-autophosphorylation step. (c) Summary of DISC1 peptide effects. MBP phosphorylation in each reaction was normalized to the reaction with Tat peptide but without pre-autophosphorylation (lane 2). (d) DISCpep-WT inhibits kinase activity of endogenous TNIK. Kinase reactions were carried out using MBP as substrate with endogenous TNIK immunoprecipitated from HEK293 cells or 22 DIV rat hippocampal cultures (top) and 10 µM of indicated peptides. The kinase input for each reaction was shown by western blotting using an anti-TNIK antibody (middle) and MBP input by Coomassie blue staining (bottom). (e) Summary of DISCpep-WTeffect on endogenous TNIK. MBP phosphorylation in each reaction was normalized to the reaction with Tat peptide and TNIK from HEK293 cells. _N_=3. *P<0.05, **P<0.01, ***P<0.001.
Figure 5
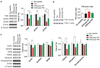
DISC1-derived peptide inhibits TNIK and regulates levels of PSD proteins in primary hippocampal neurons. (a) DISCpep-WT causes a reduction in total TNIK and phospho-TNIK levels. Primary hippocampal neurons (16–22 DIV) were treated with 10 µM of indicated peptides for 40 min. _N_= 3. (b) DISCpep-WT increases the filamentous-to-globular (F/G)-actin ratio. Cultures were treated as in (a), followed by Triton X-100 extraction to separate Triton soluble (TxS) and Triton insoluble (TxIS) fractions. TxS and TxIS contain G- and F-actin, respectively. _N_= 3. (c) DISCpep-WT causes a reduction in total levels of key PSD proteins. Peptide treatment was same as (a) except indicated proteins were analyzed. _N_=3. Total protein was normalized to β-actin, and phosphorylated protein to corresponding total protein. *P<0.05, **P<0.01, ***P<0.001.
Figure 6
Loss of PSD95 and GluR1 after inhibition of TNIK is through proteasomal and lysosomal degradation pathways. (a) Degradation of PSD-95 and GluR1 was rescued by the proteasome inhibitor, MG132, and the lysosome inhibitor, leupeptin, respectively. Primary hippocampal neurons (16–22 DIV) were pretreated with 10µM of MG132 or 25µgml−1 of leupeptin for 7 h before treatment with 10 µM of the indicated peptide for 40min. (b) Summary of the effects of MG132 and leupeptin. _N_=4. Total protein was normalized to β-actin, and phosphorylated protein to corresponding total protein. *P < 0.05, **P<0.01, ***P<0.001.
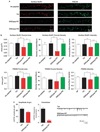
Figure 7
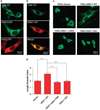
Inhibition of TNIK leads to decreases in both the surface levels of GluR1 and AMPAR mEPSCs. (a) Representative images of dendritic segments of primary hippocampal neurons (19–22 DIV) treated with 10 µM of indicated peptide for 30min, followed by immunostaining for surface GluR1 and PSD-95. (b) DISCpep-WT treatment decreases the size, density and intensity of the surface GluR1 and PSD-95 puncta. _N_= 15. (c) DISCpep-WT treatment decreases both mEPSC amplitude and frequency of primary hippocampal neurons (13 or 19 DIV) pretreated for 40 min with indicated peptides. Representative traces are shown on the right. _N_≥4. *P<0.05, **P<0.01, ***P<0.001.
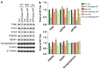
Figure 8
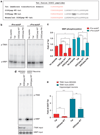
DISC1 and TNIK regulate components of the PSD in a complex fashion. (a, b) TNIK knockdown by a lentiviral-expressed shRNA regulates the level of PSD proteins in primary hippocampal cultures consistent with effects mediated by peptide inhibition. _N_=4. (c, d) TNIK shRNA knockdown causes decreases in total and surface GluR1. _N_=5. (e) TNIK shRNA knockdown causes decreases in mEPSC amplitude, but not frequency, in primary hippocampal neurons, and this decrease can be rescued by knockdown-resistant TNIK but not by WT TNIK. The representative traces are shown on the right. _N_≥4. (f) DISC1 knockdown regulates the level of PSD proteins in primary hippocampal cultures. _N_=3. Total protein was normalized to β-actin, and phosphorylated protein to corresponding total protein. *P<0.05, **P< 0.01, ***P<0.001.
Figure 8
DISC1 and TNIK regulate components of the PSD in a complex fashion. (a, b) TNIK knockdown by a lentiviral-expressed shRNA regulates the level of PSD proteins in primary hippocampal cultures consistent with effects mediated by peptide inhibition. _N_=4. (c, d) TNIK shRNA knockdown causes decreases in total and surface GluR1. _N_=5. (e) TNIK shRNA knockdown causes decreases in mEPSC amplitude, but not frequency, in primary hippocampal neurons, and this decrease can be rescued by knockdown-resistant TNIK but not by WT TNIK. The representative traces are shown on the right. _N_≥4. (f) DISC1 knockdown regulates the level of PSD proteins in primary hippocampal cultures. _N_=3. Total protein was normalized to β-actin, and phosphorylated protein to corresponding total protein. *P<0.05, **P< 0.01, ***P<0.001.
Similar articles
- Disease-associated synaptic scaffold protein CNK2 modulates PSD size and influences localisation of the regulatory kinase TNIK.
Zieger HL, Kunde SA, Rademacher N, Schmerl B, Shoichet SA. Zieger HL, et al. Sci Rep. 2020 Mar 31;10(1):5709. doi: 10.1038/s41598-020-62207-4. Sci Rep. 2020. PMID: 32235845 Free PMC article. - Caveolin-1 regulation of disrupted-in-schizophrenia-1 as a potential therapeutic target for schizophrenia.
Kassan A, Egawa J, Zhang Z, Almenar-Queralt A, Nguyen QM, Lajevardi Y, Kim K, Posadas E, Jeste DV, Roth DM, Patel PM, Patel HH, Head BP. Kassan A, et al. J Neurophysiol. 2017 Jan 1;117(1):436-444. doi: 10.1152/jn.00481.2016. Epub 2016 Nov 2. J Neurophysiol. 2017. PMID: 27832597 Free PMC article. - Neurexin-Neuroligin Synaptic Complex Regulates Schizophrenia-Related DISC1/Kal-7/Rac1 "Signalosome".
Owczarek S, Bang ML, Berezin V. Owczarek S, et al. Neural Plast. 2015;2015:167308. doi: 10.1155/2015/167308. Epub 2015 May 20. Neural Plast. 2015. PMID: 26078884 Free PMC article. - DISC1-binding proteins in neural development, signalling and schizophrenia.
Bradshaw NJ, Porteous DJ. Bradshaw NJ, et al. Neuropharmacology. 2012 Mar;62(3):1230-41. doi: 10.1016/j.neuropharm.2010.12.027. Epub 2010 Dec 31. Neuropharmacology. 2012. PMID: 21195721 Free PMC article. Review. - Mitochondrial roles of the psychiatric disease risk factor DISC1.
Norkett R, Modi S, Kittler JT. Norkett R, et al. Schizophr Res. 2017 Sep;187:47-54. doi: 10.1016/j.schres.2016.12.025. Epub 2017 Jan 10. Schizophr Res. 2017. PMID: 28087269 Review.
Cited by
- DISC1 Pathway in Brain Development: Exploring Therapeutic Targets for Major Psychiatric Disorders.
Kamiya A, Sedlak TW, Pletnikov MV. Kamiya A, et al. Front Psychiatry. 2012 Mar 22;3:25. doi: 10.3389/fpsyt.2012.00025. eCollection 2012. Front Psychiatry. 2012. PMID: 22461775 Free PMC article. - The TRAX, DISC1, and GSK3 complex in mental disorders and therapeutic interventions.
Weng YT, Chien T, Kuan II, Chern Y. Weng YT, et al. J Biomed Sci. 2018 Oct 4;25(1):71. doi: 10.1186/s12929-018-0473-x. J Biomed Sci. 2018. PMID: 30285728 Free PMC article. Review. - Misassembly of full-length Disrupted-in-Schizophrenia 1 protein is linked to altered dopamine homeostasis and behavioral deficits.
Trossbach SV, Bader V, Hecher L, Pum ME, Masoud ST, Prikulis I, Schäble S, de Souza Silva MA, Su P, Boulat B, Chwiesko C, Poschmann G, Stühler K, Lohr KM, Stout KA, Oskamp A, Godsave SF, Müller-Schiffmann A, Bilzer T, Steiner H, Peters PJ, Bauer A, Sauvage M, Ramsey AJ, Miller GW, Liu F, Seeman P, Brandon NJ, Huston JP, Korth C. Trossbach SV, et al. Mol Psychiatry. 2016 Nov;21(11):1561-1572. doi: 10.1038/mp.2015.194. Epub 2016 Jan 12. Mol Psychiatry. 2016. PMID: 26754951 Free PMC article. - DISC1: Structure, Function, and Therapeutic Potential for Major Mental Illness.
Soares DC, Carlyle BC, Bradshaw NJ, Porteous DJ. Soares DC, et al. ACS Chem Neurosci. 2011 Nov 16;2(11):609-632. doi: 10.1021/cn200062k. Epub 2011 Aug 5. ACS Chem Neurosci. 2011. PMID: 22116789 Free PMC article. - Organization of TNIK in dendritic spines.
Burette AC, Phend KD, Burette S, Lin Q, Liang M, Foltz G, Taylor N, Wang Q, Brandon NJ, Bates B, Ehlers MD, Weinberg RJ. Burette AC, et al. J Comp Neurol. 2015 Sep 1;523(13):1913-24. doi: 10.1002/cne.23770. Epub 2015 Jul 1. J Comp Neurol. 2015. PMID: 25753355 Free PMC article.
References
- Sodhi M, Wood KH, Meador-Woodruff J. Role of glutamate in schizophrenia: integrating excitatory avenues of research. Expert Rev Neurother. 2008;8:1389–1406. - PubMed
- Coyle JT. Glutamate and schizophrenia: beyond the dopamine hypothesis. Cell Mol Neurobiol. 2006;26:365–384. - PubMed
- Schiffer HH. Glutamate receptor genes: susceptibility factors in schizophrenia and depressive disorders? Mol Neurobiol. 2002;25:191–212. - PubMed
MeSH terms
Substances
Grants and funding
- R01 NS035527-11/NS/NINDS NIH HHS/United States
- R01 NS035527-12/NS/NINDS NIH HHS/United States
- R01 NS035527-08A1/NS/NINDS NIH HHS/United States
- R01 NS035527-10/NS/NINDS NIH HHS/United States
- R01 NS035527-09/NS/NINDS NIH HHS/United States
- G0600765/MRC_/Medical Research Council/United Kingdom
- R01 NS035527/NS/NINDS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials