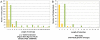The characterization of twenty sequenced human genomes - PubMed (original) (raw)
. 2010 Sep 9;6(9):e1001111.
doi: 10.1371/journal.pgen.1001111.
Kevin V Shianna, Dongliang Ge, Jessica M Maia, Mingfu Zhu, Jason P Smith, Elizabeth T Cirulli, Jacques Fellay, Samuel P Dickson, Curtis E Gumbs, Erin L Heinzen, Anna C Need, Elizabeth K Ruzzo, Abanish Singh, C Ryan Campbell, Linda K Hong, Katharina A Lornsen, Alexander M McKenzie, Nara L M Sobreira, Julie E Hoover-Fong, Joshua D Milner, Ruth Ottman, Barton F Haynes, James J Goedert, David B Goldstein
Affiliations
- PMID: 20838461
- PMCID: PMC2936541
- DOI: 10.1371/journal.pgen.1001111
The characterization of twenty sequenced human genomes
Kimberly Pelak et al. PLoS Genet. 2010.
Abstract
We present the analysis of twenty human genomes to evaluate the prospects for identifying rare functional variants that contribute to a phenotype of interest. We sequenced at high coverage ten "case" genomes from individuals with severe hemophilia A and ten "control" genomes. We summarize the number of genetic variants emerging from a study of this magnitude, and provide a proof of concept for the identification of rare and highly-penetrant functional variants by confirming that the cause of hemophilia A is easily recognizable in this data set. We also show that the number of novel single nucleotide variants (SNVs) discovered per genome seems to stabilize at about 144,000 new variants per genome, after the first 15 individuals have been sequenced. Finally, we find that, on average, each genome carries 165 homozygous protein-truncating or stop loss variants in genes representing a diverse set of pathways.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
Figure 1. Average per-genome overlap between SNVs in genomic databases and SNVs identified by whole-genome sequencing.
On average, 3,473,639 SNVs were observed in each genome (Table S2). A per-genome average of 87.28% of these SNVs were present in the dbSNP database (version 129, validated) (Table S3).
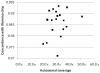
Figure 2. Concordance between sequencing and genotyping calls.
The sequenced samples were also run on either the Illumina Human 1M-Duo v3 BeadChip or the Illumina 610-Quad BeadChip. The concordance rate between the sequencing and the Illumina BeadChip genotype calls is plotted against sequencing coverage of the autosomes. A data point is plotted for each of the twenty genomes.
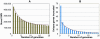
Figure 3. Coding indel length distribution.
Shown is a side-by-side comparison of the length of the coding indels in this study as compared to a previous publication . (A) Indel lengths observed in J.C. Venter's exome versus (B) indel lengths observed in this study. The data from our study have been restricted to the canonical genes or transcripts that are captured by the Agilent SureSelect Targeted Enrichment system. Indels that are a multiple of 3bp in length are marked in green.
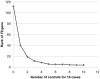
Figure 4. Rank of the F8 gene as the number of control genomes increases.
The gene ranking was ordered by the number of case genomes that carried protein-truncating or stop loss variants, in homozygous form or on the X-chromosome, that were not present in control genomes in homozygous form. Ranking was performed with a “gene prioritization” function implemented in the SVA software tool (Text S1). Protein-truncating variants were defined as SNVs that cause a premature stop codon, and insertions or deletions that cause a frameshift coding change. The ranks represent an average taken from five permutations. When comparing 10 hemophilia cases to just one control, F8 ranks in the top 40 genes. Once 5 or more controls are available, it ranks in the top 5 genes.
Figure 5. Number of novel SNVs and novel knocked-out genes as the number of genomes increases.
The total number of novel variants, and the total number of novel genes containing protein truncating or stop loss variants, continues to drop as additional genomes are added to the analysis. Shown are the number of unique SNVs (A) and unique genes carrying a homozygous protein-truncating or stop loss variant (B) per genome, as a function of the number of genomes already considered. The genomes were added in a random order to both analyses, and 1000 permutations were performed and averaged.
Similar articles
- Comprehensive characterization of human genome variation by high coverage whole-genome sequencing of forty four Caucasians.
Shen H, Li J, Zhang J, Xu C, Jiang Y, Wu Z, Zhao F, Liao L, Chen J, Lin Y, Tian Q, Papasian CJ, Deng HW. Shen H, et al. PLoS One. 2013;8(4):e59494. doi: 10.1371/journal.pone.0059494. Epub 2013 Apr 5. PLoS One. 2013. PMID: 23577066 Free PMC article. - Targeted capture and massively parallel sequencing of 12 human exomes.
Ng SB, Turner EH, Robertson PD, Flygare SD, Bigham AW, Lee C, Shaffer T, Wong M, Bhattacharjee A, Eichler EE, Bamshad M, Nickerson DA, Shendure J. Ng SB, et al. Nature. 2009 Sep 10;461(7261):272-6. doi: 10.1038/nature08250. Epub 2009 Aug 16. Nature. 2009. PMID: 19684571 Free PMC article. - KoVariome: Korean National Standard Reference Variome database of whole genomes with comprehensive SNV, indel, CNV, and SV analyses.
Kim J, Weber JA, Jho S, Jang J, Jun J, Cho YS, Kim HM, Kim H, Kim Y, Chung O, Kim CG, Lee H, Kim BC, Han K, Koh I, Chae KS, Lee S, Edwards JS, Bhak J. Kim J, et al. Sci Rep. 2018 Apr 4;8(1):5677. doi: 10.1038/s41598-018-23837-x. Sci Rep. 2018. PMID: 29618732 Free PMC article. - The Canadian "National Program for hemophilia mutation testing" database: a ten-year review.
Rydz N, Leggo J, Tinlin S, James P, Lillicrap D. Rydz N, et al. Am J Hematol. 2013 Dec;88(12):1030-4. doi: 10.1002/ajh.23557. Epub 2013 Sep 9. Am J Hematol. 2013. PMID: 23913812 Review. - Copy Number Variation and Risk of Stroke.
Grond-Ginsbach C, Erhart P, Chen B, Kloss M, Engelter ST, Cole JW. Grond-Ginsbach C, et al. Stroke. 2018 Oct;49(10):2549-2554. doi: 10.1161/STROKEAHA.118.020371. Stroke. 2018. PMID: 30355123 Free PMC article. Review. No abstract available.
Cited by
- A Comprehensive Analysis of 3 Moroccan Genomes Revealed Contributions From Both African and European Ancestries.
Boumajdi N, Bendani H, Kartti S, Alouane T, Belyamani L, Ibrahimi A. Boumajdi N, et al. Evol Bioinform Online. 2024 Feb 6;20:11769343241229278. doi: 10.1177/11769343241229278. eCollection 2024. Evol Bioinform Online. 2024. PMID: 38327511 Free PMC article. - Molecular genetic testing and the future of clinical genomics.
Katsanis SH, Katsanis N. Katsanis SH, et al. Nat Rev Genet. 2013 Jun;14(6):415-26. doi: 10.1038/nrg3493. Nat Rev Genet. 2013. PMID: 23681062 Free PMC article. Review. - SVA: software for annotating and visualizing sequenced human genomes.
Ge D, Ruzzo EK, Shianna KV, He M, Pelak K, Heinzen EL, Need AC, Cirulli ET, Maia JM, Dickson SP, Zhu M, Singh A, Allen AS, Goldstein DB. Ge D, et al. Bioinformatics. 2011 Jul 15;27(14):1998-2000. doi: 10.1093/bioinformatics/btr317. Epub 2011 May 29. Bioinformatics. 2011. PMID: 21624899 Free PMC article. - Genomics really gets personal: how exome and whole genome sequencing challenge the ethical framework of human genetics research.
Tabor HK, Berkman BE, Hull SC, Bamshad MJ. Tabor HK, et al. Am J Med Genet A. 2011 Dec;155A(12):2916-24. doi: 10.1002/ajmg.a.34357. Epub 2011 Oct 28. Am J Med Genet A. 2011. PMID: 22038764 Free PMC article. - Global assessment of genomic variation in cattle by genome resequencing and high-throughput genotyping.
Zhan B, Fadista J, Thomsen B, Hedegaard J, Panitz F, Bendixen C. Zhan B, et al. BMC Genomics. 2011 Nov 14;12:557. doi: 10.1186/1471-2164-12-557. BMC Genomics. 2011. PMID: 22082336 Free PMC article.
References
- Wheeler DA, Srinivasan M, Egholm M, Shen Y, Chen L, et al. The complete genome of an individual by massively parallel DNA sequencing. Nature. 2008;452:872–876. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- U01 AI067854/AI/NIAID NIH HHS/United States
- AI067854/AI/NIAID NIH HHS/United States
- U19 AI067854/AI/NIAID NIH HHS/United States
- RC2 NS070344/NS/NINDS NIH HHS/United States
- RC2 MH089915/MH/NIMH NIH HHS/United States
- RC2MH089915/MH/NIMH NIH HHS/United States
- RC2NS070344/NS/NINDS NIH HHS/United States
- ImNIH/Intramural NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources