Elucidation of seventeen human peripheral blood B-cell subsets and quantification of the tetanus response using a density-based method for the automated identification of cell populations in multidimensional flow cytometry data - PubMed (original) (raw)
. 2010;78 Suppl 1(Suppl 1):S69-82.
doi: 10.1002/cyto.b.20554.
Chungwen Wei, F Eun-Hyung Lee, John Campbell, Jessica Halliley, Jamie A Lee, Jennifer Cai, Y Megan Kong, Eva Sadat, Elizabeth Thomson, Patrick Dunn, Adam C Seegmiller, Nitin J Karandikar, Christopher M Tipton, Tim Mosmann, Iñaki Sanz, Richard H Scheuermann
Affiliations
- PMID: 20839340
- PMCID: PMC3084630
- DOI: 10.1002/cyto.b.20554
Elucidation of seventeen human peripheral blood B-cell subsets and quantification of the tetanus response using a density-based method for the automated identification of cell populations in multidimensional flow cytometry data
Yu Qian et al. Cytometry B Clin Cytom. 2010.
Abstract
Background: Advances in multiparameter flow cytometry (FCM) now allow for the independent detection of larger numbers of fluorochromes on individual cells, generating data with increasingly higher dimensionality. The increased complexity of these data has made it difficult to identify cell populations from high-dimensional FCM data using traditional manual gating strategies based on single-color or two-color displays.
Methods: To address this challenge, we developed a novel program, FLOCK (FLOw Clustering without K), that uses a density-based clustering approach to algorithmically identify biologically relevant cell populations from multiple samples in an unbiased fashion, thereby eliminating operator-dependent variability.
Results: FLOCK was used to objectively identify seventeen distinct B-cell subsets in a human peripheral blood sample and to identify and quantify novel plasmablast subsets responding transiently to tetanus and other vaccinations in peripheral blood. FLOCK has been implemented in the publically available Immunology Database and Analysis Portal-ImmPort (http://www.immport.org)-for open use by the immunology research community.
Conclusions: FLOCK is able to identify cell subsets in experiments that use multiparameter FCM through an objective, automated computational approach. The use of algorithms like FLOCK for FCM data analysis obviates the need for subjective and labor-intensive manual gating to identify and quantify cell subsets. Novel populations identified by these computational approaches can serve as hypotheses for further experimental study.
© 2010 International Clinical Cytometry Society.
Figures
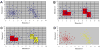
Figure 1. Algorithmic components of FLOCK
The FLOCKalgorithm consists of four core steps: hyper-grid creation, identifying dense hyper-regions, merging neighboring dense hyper-regions, clustering based on centroids derived from the merged dense hyper-regions. A) A gridding example. Each dimension corresponds to one measured characteristics (e.g. fluorescence at a given wavelength). Each dot is one event (e.g. data about a cell) with characteristic values (e.g. fluorescence intensity) as its coordinates. A number of equal-sized partitions (in this example the number is ten) are created on each dimension to generate a hyper-grid. B) Dense data hyper-regions. Density of a data hyper-region is calculated based on the number of event inside the region. A density threshold is determined by a data-driven approach described in the Methods section. Hyper-regions having larger number of events than the density threshold are considered dense. In this example, the density threshold is set at two and the resulting dense hyper-regions are marked in red. C) Merging dense hyper-regions. Dense hyper-regions are merged if they neighbor each other. The number of merged hyper-region groups (in this example the number is two, highlighted in red and yellow) is used as the number of final clusters, and the centroid of the events in a given hyper-region group calculated. D) Final clustering. Dots (events) are assigned to the closest centroid of the merged dense region group based on Euclidean distance to form the two populations.
Figure 2. Analysis of B cell populations in normal blood using FLOCK
Following file conversion, data for IgD, IgG, CD24, B220, CD38 and CD27 fluorescence intensity was extracted and used for FLOCK analysis with automated parameter selection. Data has been CD3-CD19+ pre-gated. A) Two-dimensional dot plot displays for IgD vs. CD27, CD24 vs B220, and IgG vs CD38. The first row shows FlowJo displays of selected two-dimensional dot plots with manual gating applied to the IgD vs CD27 display; gated population proportions are provided. FLOCK-identified populations are displayed in the second row with each of the FLOCK-identified cell subsets highlighted in a different color. The six regions of the IgD vs CD27 dot plot corresponding to previously defined naïve and memory B cell subsets are highlighted - N (naïve), UM (unswitched memory), GSM (IgG positive switched memory), GNSM (IgG negative switched memory), (DNM (double negative memory), and PB (plasmablasts)). CD24 vs B220 and IgG vs CD38 dot plots showing individual populations for each of the naïve (B), unswitched memory (C), IgG positive switched memory (D), IgG negative switched memory (E), double negative memory (F), and plasmablasts (G). The IgD vs CD27 plots for the individual populations are included in Supplemental Figure 1.
Figure 3. Analysis of B cell responses to tetanus vaccination
B cell subsets identified using CD19, CD27, IgD, CD38, CD138, CXCR4, and HLA-DR expression from peripheral blood of a subject pre (day 0) and post tetanus vaccination (Day 5, 6, 7, 8 and 15). (A) Twelve populations identified by FLOCK analysis as shown by CD27 and IgD plots of the Day 6 sample, and further characterized in Supplemental Figure 1. (B) Three CD19+ plasmablast-like subsets were identified - Pop5: CD138−, CD38+, Ki67+, Pop6: CD138−, CD38hi, Ki67+, and Pop7: CD138+, CD38hi, Ki67+. (C, D) Total IgG and tetanus-specific IgG antibody-secreting cells (ASC) were identified by Elispot assay, and Pop 6 and 7 plasmablast-like subsets were identified by FLOCK and manual gating, respectively. (E) Pax5 and BLIMP-1 mRNA expression in Populations #5, #6, and #7.
Figure 4. Reproducible detection of plasmablast/plasma cell populations in vaccination subjects using FLOCK
The centroid locations for the B cell subsets identified in the tetanus study (Figure 3) were used to identify the plasmablast (Pop5 & Pop6) and plasma cell (Pop7) subsets in six independent subjects vaccinated with the indicated vaccines – tetanus and diptheria toxoids (Tet), trivalent influenza vaccine 2009 (TIV), H1N1 monovalent influenza vaccine 2009 (H1N1), Hepatitis A(HepA), and Hepatitis B (HepB). (A) CD38 versus CD138 dot plots with individual cell events for the specific cell populations color-coded. Percentages of population events in the CD19+CD3- gated parent population are indicated. (B) Population percentages determined using FLOCK and manual gating are compared. Pearson two-tailed correlation coefficient (r) was used to assess the degree of correlation.
Similar articles
- DAFi: A directed recursive data filtering and clustering approach for improving and interpreting data clustering identification of cell populations from polychromatic flow cytometry data.
Lee AJ, Chang I, Burel JG, Lindestam Arlehamn CS, Mandava A, Weiskopf D, Peters B, Sette A, Scheuermann RH, Qian Y. Lee AJ, et al. Cytometry A. 2018 Jun;93(6):597-610. doi: 10.1002/cyto.a.23371. Epub 2018 Apr 17. Cytometry A. 2018. PMID: 29665244 Free PMC article. - Validation of a hybrid approach to standardize immunophenotyping analysis in large population studies: The Health and Retirement Study.
Hunter-Schlichting D, Lane J, Cole B, Flaten Z, Barcelo H, Ramasubramanian R, Cassidy E, Faul J, Crimmins E, Pankratz N, Thyagarajan B. Hunter-Schlichting D, et al. Sci Rep. 2020 May 29;10(1):8759. doi: 10.1038/s41598-020-65016-x. Sci Rep. 2020. PMID: 32472068 Free PMC article. - Flow cytometric immunophenotyping of lymphocyte subsets in samples that contain a high proportion of non-lymphoid cells.
Pattanapanyasat K, Kyle DE, Tongtawe P, Yongvanitchit K, Fucharoen S. Pattanapanyasat K, et al. Cytometry. 1994 Dec 15;18(4):199-208. doi: 10.1002/cyto.990180403. Cytometry. 1994. PMID: 7534676 - Automated Analysis of Clinical Flow Cytometry Data: A Chronic Lymphocytic Leukemia Illustration.
Scheuermann RH, Bui J, Wang HY, Qian Y. Scheuermann RH, et al. Clin Lab Med. 2017 Dec;37(4):931-944. doi: 10.1016/j.cll.2017.07.011. Clin Lab Med. 2017. PMID: 29128077 Free PMC article. Review. - Assays for B cell and germinal center development.
Bleesing JJH. Bleesing JJH. Curr Protoc Immunol. 2004 Nov;Chapter 7:7.35.1-7.35.21. doi: 10.1002/0471142735.im0735s63. Curr Protoc Immunol. 2004. PMID: 18432934 Review.
Cited by
- Association between magnitude of the virus-specific plasmablast response and disease severity in dengue patients.
Garcia-Bates TM, Cordeiro MT, Nascimento EJ, Smith AP, Soares de Melo KM, McBurney SP, Evans JD, Marques ET Jr, Barratt-Boyes SM. Garcia-Bates TM, et al. J Immunol. 2013 Jan 1;190(1):80-7. doi: 10.4049/jimmunol.1103350. Epub 2012 Nov 30. J Immunol. 2013. PMID: 23203929 Free PMC article. - Immunophenotype Discovery, Hierarchical Organization, and Template-Based Classification of Flow Cytometry Samples.
Azad A, Rajwa B, Pothen A. Azad A, et al. Front Oncol. 2016 Aug 31;6:188. doi: 10.3389/fonc.2016.00188. eCollection 2016. Front Oncol. 2016. PMID: 27630823 Free PMC article. - Inferring phenotypic properties from single-cell characteristics.
Qiu P. Qiu P. PLoS One. 2012;7(5):e37038. doi: 10.1371/journal.pone.0037038. Epub 2012 May 25. PLoS One. 2012. PMID: 22662133 Free PMC article. - Differences in B-Cell Immunophenotypes and Neutralizing Antibodies Against SARS-CoV-2 After Administration of BNT162b2 (Pfizer-BioNTech) Vaccine in Individuals with and without Prior COVID-19 - A Prospective Cohort Study.
Morales-Núñez JJ, García-Chagollán M, Muñoz-Valle JF, Díaz-Pérez SA, Torres-Hernández PC, Rodríguez-Reyes SC, Santoscoy-Ascencio G, Sierra García de Quevedo JJ, Hernández-Bello J. Morales-Núñez JJ, et al. J Inflamm Res. 2022 Aug 4;15:4449-4466. doi: 10.2147/JIR.S374304. eCollection 2022. J Inflamm Res. 2022. PMID: 35958186 Free PMC article. - Understanding B-cell activation and autoantibody repertoire selection in systemic lupus erythematosus: A B-cell immunomics approach.
Tipton CM, Hom JR, Fucile CF, Rosenberg AF, Sanz I. Tipton CM, et al. Immunol Rev. 2018 Jul;284(1):120-131. doi: 10.1111/imr.12660. Immunol Rev. 2018. PMID: 29944759 Free PMC article. Review.
References
- Agematsu K, Nagumo H, Yang F, Nakazawa T, Fukushima K, Ito S, Sugita K, Mori T, Kobata T, Morimoto C, Komiyama A. B cell subpopulations separated by CD27 and crucial collaboration of CD27+ B cells and helper T cells in immunoglobulin production. European Journal of Immunology. 1997;27:2073–2079. - PubMed
- Agrawal R, Gehrke J, Gunopulos D, Raghavan P. Automated subspace clustering of high-dimensional data for data mining applications. Proceedings of SIGMOD Conference on Management of Data; 1998. pp. 94–105.
- Al-Mawali A, Gillis D, Lewis I. The role of multiparameter flow cytometry for detection of minimal residual disease in acute myeloid leukemia. American Journal of Clinical Pathology. 2009;131:16–26. - PubMed
- Arce S, Luger E, Muehlinghaus G, Cassese G, Hauser A, Horst A, Lehnert K, Odendahl M, Hönemann D, Heller K-D, Kleinschmidt H, Berek C, Dörner T, Krenn V, Hiepe F, Bargou R, Radbruch A, Manz RA. CD38 low IgG-secreting cells are precursors of various CD38 high-expressing plasma cell populations. Journal of Leukocyte Biology. 2004;75:1022–1028. - PubMed
- Aurran-Schleinitz T, Telford W, Perfetto S, Caporaso N, Wilson W, Stetler-Stevenson MA, Zenger VE, Abbasi F, Marti GE. Identification of a new monoclonal B-cell subset in unaffected first-degree relatives in familial chronic lymphocytic leukemia. Leukemia. 2005;19:2339–2341. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- N01AI50029/AI/NIAID NIH HHS/United States
- R37 AI049660/AI/NIAID NIH HHS/United States
- N01 AI50029/AI/NIAID NIH HHS/United States
- U19-AI56390/AI/NIAID NIH HHS/United States
- U19 AI056390-08/AI/NIAID NIH HHS/United States
- K24 AI079272/AI/NIAID NIH HHS/United States
- U19 AI056390/AI/NIAID NIH HHS/United States
- N01 AI040076/AI/NIAID NIH HHS/United States
- N01AI40076/AI/NIAID NIH HHS/United States
- N01-AI50029/AI/NIAID NIH HHS/United States
- N01-AI40076/AI/NIAID NIH HHS/United States
- R01 AI084808/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical