A role for the dynamin-like protein Vps1 during endocytosis in yeast - PubMed (original) (raw)
. 2010 Oct 15;123(Pt 20):3496-506.
doi: 10.1242/jcs.070508. Epub 2010 Sep 14.
Affiliations
- PMID: 20841380
- PMCID: PMC2951468
- DOI: 10.1242/jcs.070508
A role for the dynamin-like protein Vps1 during endocytosis in yeast
Iwona I Smaczynska-de Rooij et al. J Cell Sci. 2010.
Abstract
Dynamins are a conserved family of proteins involved in membrane fusion and fission. Although mammalian dynamins are known to be involved in several membrane-trafficking events, the role of dynamin-1 in endocytosis is the best-characterised role of this protein family. Despite many similarities between endocytosis in yeast and mammalian cells, a comparable role for dynamins in yeast has not previously been demonstrated. The reported lack of involvement of dynamins in yeast endocytosis has raised questions over the general applicability of the current yeast model of endocytosis, and has also precluded studies using well-developed methods in yeast, to further our understanding of the mechanism of dynamin function during endocytosis. Here, we investigate the yeast dynamin-like protein Vps1 and demonstrate a transient burst of localisation to sites of endocytosis. Using live-cell imaging of endocytic reporters in strains lacking vps1, and also electron microscopy and biochemical approaches, we demonstrate a role for Vps1 in facilitating endocytic invagination. Vps1 mutants were generated, and analysis in several assays reveals a role for the C-terminal self-assembly domain in endocytosis but not in other membrane fission events with which Vps1 has previously been associated.
Figures
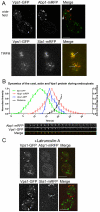
Fig. 1.
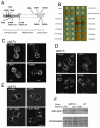
Vps1 colocalises with endocytic proteins. (A) Vps1-GFP and either Abp1-mRFP (upper) or Sla1-mRFP (lower) were coexpressed and cells imaged. Arrows depict merged spots. Scale bar: 2 μm. (B) Vps1-GFP was coexpressed with Abp1-mRFP and intensity curves generated for eight separate endocytic spots. Weighted averages of spot intensity was calculated and plotted (means ± s.d.). The accompanying images for one of these spots are shown below the graph. Sla1-GFP was also expressed with Abp1-mRFP and intensity curves generated to allow relative comparisons. The distance of the tip of the Abp1 spots from the membrane were plotted. (C) 200 μM Lat-A was added to cells coexpressing Vps1-GFP with Abp1-mRFP (upper) or Sla1-mRFP (lower). Scale bars: 2 μm.
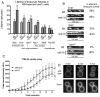
Fig. 2.
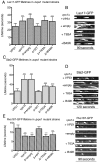
The effect of vps1 deletion on known endocytic proteins. (A) The lifetime of six endocytic proteins, Sla2, Ent1, Las17, Abp1, Sac6, and Rvs167 was analysed in the presence and absence of vps1. For wild-type cells, _n_=67, 22,119, 30, 60, 35 and for _vps1_Δ cells, _n_=19, 36, 121, 36, 69, 33 for the markers as ordered on graph. Error bars indicate s.d. ***P<0.0001 for Sla2, Ent1, Las17, Abp1; **P<0.0004 for Sac6 and 0.0002 for Rvs167. (B) Kymographs of Las17-GFP, Sla2-GFP and Ent1-GFP. Aberrant endocytic events for Sla2 and Ent1 were defined as those showing no invagination, movement in the membrane plane, delayed scission or retraction. For Las17, aberrant events were those showing movement in the membrane plane, inappropriate invagination and retraction. (C) FM4-64 internalisation was performed as described. Total internal cell fluorescence intensity (mean ± s.d.) was measured at each time point. (D) Representative images of wild-type and _vps1_Δ cells from three time points in the FM4-64-uptake assay. Arrows indicate foci of FM4-64 staining on the plasma membrane. Scale bars: 2 μm.
Fig. 3.
The effect of vps1 deletion on the amphiphysin Rvs167. (A) Strains carrying single deletions of either _vps1_Δ or _rvs167_Δ or a double-deletion strain were assessed for growth at 30°C and 37°C. (B) Rvs167-GFP localised in wild-type and _vps1_Δ cells. Images were recorded for 500 mseconds. Shown is a representative field of cells. Scale bars: 5 μm. (C) Fluorescence intensity of Rvs167-GFP patches and cytoplasmic background in wild-type and _vps1_Δ cells were recorded and plotted. Mean patch intensity: wild type, 885±508 units, _n_=128; vps1Δ, 429±227 units, _n_=53; P<0.0001. (D) Vps1-GFP was coexpressed with Rvs167-mRFP and intensity curves generated. Weighted averages of spot intensity were calculated and plotted. (E) Kymographs of Sac6-mRFP in wild-type, _vps1_Δ, _rvs167_Δ cells and in _vps1_Δ_rvs167_Δ cells. Percentage of spots that retracted toward the membrane was counted. _n_=210, wild type; 46, _vps1_Δ; 52, _rvs167_Δ; 48, _vps1_Δ_rvs167_Δ cells.
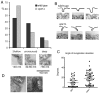
Fig. 4.
Ultrastructural analysis of endocytic patches in _vps1_Δ cells. (A) The proportion of shallow, pronounced and deep invaginations in wild-type and _vps1_Δ cells was counted (_n_=151, wild type; _n_=91, _vps1_Δ). (B) Images of pronounced invaginations in cells in the presence and absence of Vps1. Above the micrographs are outline tracings showing the invagination profile. (C) The angle of invagination relative to a line perpendicular to the plasma membrane was measured for both wild-type and _vps1_Δ invaginations. Horizontal lines indicate the means ± s.d. (D) In pronounced invaginations in wild-type cells, a repeated structure at the side of the invaginations can be observed in sections (arrows).
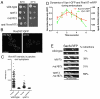
Fig. 5.
Analysis of the endosome, vacuole and peroxisome in Vps1 mutants. (A) Mutations were generated in Vps1 as depicted. (B) Cells deleted for vps1 were transformed with a construct expressing the recycling SNARE protein Snc1, fused to SUC2 and GFP. Localisation of invertase is detected in a colorimetric assay. (C) Localisation of GFP-Snc1-SUC2 construct was analysed in wild-type, _vps1_Δ, T63A and I649K mutants. Scale bars: 2 μm. (D) Uptake of FM4-64 reveals vacuolar trafficking defects in a subset of vps1 mutant strains. Cells were incubated for 10 minutes with FM4-64, washed and then visualised after 90 minutes. Arrows indicate clusters of endosomes or fragmented vacuoles surrounding a large weakly stained vacuole, the class F phenotype. Scale bars: 10 μm. (E) Peroxisomes labelled with GFP peroxisome reporter were visualised in a _vps1_Δ_dnm1_Δ strain expressing plasmids with mutant versions of vps1. Images are compressed _Z_-stacks for fission-proficient strains and in a single plane for fission-deficient strains. Scale bar: 5 μm. (F) Whole-cell extracts separated by SDS–PAGE. The upper part of gel was used for western blotting with mouse anti-Vps1 polyclonal antibody. The lower half of the gel was removed and stained with Coomassie Blue to indicate protein-loading levels.
Fig. 6.
Vps1 mutants show defects in the behaviour of endocytic proteins. (A) Lifetime of Las17-GFP was measured in wild-type and _vps1_Δ cells, and in _vps1_Δ strains transformed with plasmids (empty, VPS1, vps1T63A and Vps1I649K). _n_=119, 121, 24, 34, 41, 25, respectively. Error bars indicate s.d. All mutant vps1 lifetimes are significantly different from the wild type (***P<0.0001). (B) Kymographs of Las17-GFP in the strains. (C) Lifetime of Sla2-GFP was measured in wild-type and _vps1_Δ cells and in _vps1_Δ strains transformed with the plasmids as indicated. _n_=67, 24, 24, 35, 40, 23, respectively. Error bars indicate s.d. All mutants are significantly different from the wild-type and null strains (***P<0.0001). (D) Kymographs of Sla2-GFP in each of these mutants. Exposure time was increased to allow visualisation of spots in the mutant and null strains. (E) Lifetime of Rvs167-GFP in wild-type and _vps1_Δ cells and in _vps1_Δ strains transformed with the plasmids as indicated. _n_=35, 33, 116, 91, 114, 108, respectively. Error bars indicate s.d. All mutants are significantly different from wild type (***P<0.0001).
Fig. 7.
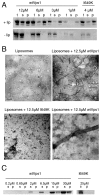
Vps1 binds and tubulates liposomes. (A) Vps1 was incubated with liposomes, which were then subjected to centrifugation and total protein (t), pellets (p) and supernatants (s) were analysed by SDS–PAGE. Varying concentrations of wtVps1, or 4 μM I649K Vps1 were used in the presence (top panel) or absence (bottom panel) of liposomes. (B) Liposomes were analysed using electron microscopy in the presence and absence of wt Vps1 or I649K Vps1. A high-resolution image of a tubulated liposome reveals a regular organisation of negative staining along the formed tubule. (C) Self-assembly assays of wt Vps1 and I649K Vps1 were performed as described. s, supernatant; p, pellet fractions.
Similar articles
- Mutation of key lysine residues in the Insert B region of the yeast dynamin Vps1 disrupts lipid binding and causes defects in endocytosis.
Smaczynska-de Rooij II, Marklew CJ, Palmer SE, Allwood EG, Ayscough KR. Smaczynska-de Rooij II, et al. PLoS One. 2019 Apr 22;14(4):e0215102. doi: 10.1371/journal.pone.0215102. eCollection 2019. PLoS One. 2019. PMID: 31009484 Free PMC article. - Yeast dynamin Vps1 and amphiphysin Rvs167 function together during endocytosis.
Smaczynska-de Rooij II, Allwood EG, Mishra R, Booth WI, Aghamohammadzadeh S, Goldberg MW, Ayscough KR. Smaczynska-de Rooij II, et al. Traffic. 2012 Feb;13(2):317-28. doi: 10.1111/j.1600-0854.2011.01311.x. Epub 2011 Dec 6. Traffic. 2012. PMID: 22082017 - Phosphorylation Regulates the Endocytic Function of the Yeast Dynamin-Related Protein Vps1.
Smaczynska-de Rooij II, Marklew CJ, Allwood EG, Palmer SE, Booth WI, Mishra R, Goldberg MW, Ayscough KR. Smaczynska-de Rooij II, et al. Mol Cell Biol. 2015 Dec 28;36(5):742-55. doi: 10.1128/MCB.00833-15. Mol Cell Biol. 2015. PMID: 26711254 Free PMC article. - From membranes to organelles: emerging roles for dynamin-like proteins in diverse cellular processes.
Williams M, Kim K. Williams M, et al. Eur J Cell Biol. 2014 Jul;93(7):267-77. doi: 10.1016/j.ejcb.2014.05.002. Epub 2014 Jun 2. Eur J Cell Biol. 2014. PMID: 24954468 Review. - The VPS1 protein is a dynamin-like GTPase required for sorting proteins to the yeast vacuole.
Ekena K, Vater CA, Raymond CK, Stevens TH. Ekena K, et al. Ciba Found Symp. 1993;176:198-211; discussion 211-4. doi: 10.1002/9780470514450.ch13. Ciba Found Symp. 1993. PMID: 8299420 Review.
Cited by
- Structures of the fungal dynamin-related protein Vps1 reveal a unique, open helical architecture.
Varlakhanova NV, Alvarez FJD, Brady TM, Tornabene BA, Hosford CJ, Chappie JS, Zhang P, Ford MGJ. Varlakhanova NV, et al. J Cell Biol. 2018 Oct 1;217(10):3608-3624. doi: 10.1083/jcb.201712021. Epub 2018 Aug 7. J Cell Biol. 2018. PMID: 30087125 Free PMC article. - Mutation of key lysine residues in the Insert B region of the yeast dynamin Vps1 disrupts lipid binding and causes defects in endocytosis.
Smaczynska-de Rooij II, Marklew CJ, Palmer SE, Allwood EG, Ayscough KR. Smaczynska-de Rooij II, et al. PLoS One. 2019 Apr 22;14(4):e0215102. doi: 10.1371/journal.pone.0215102. eCollection 2019. PLoS One. 2019. PMID: 31009484 Free PMC article. - An Abp1-dependent route of endocytosis functions when the classical endocytic pathway in yeast is inhibited.
Aghamohammadzadeh S, Smaczynska-de Rooij II, Ayscough KR. Aghamohammadzadeh S, et al. PLoS One. 2014 Jul 29;9(7):e103311. doi: 10.1371/journal.pone.0103311. eCollection 2014. PLoS One. 2014. PMID: 25072293 Free PMC article. - Membrane scission driven by the PROPPIN Atg18.
Gopaldass N, Fauvet B, Lashuel H, Roux A, Mayer A. Gopaldass N, et al. EMBO J. 2017 Nov 15;36(22):3274-3291. doi: 10.15252/embj.201796859. Epub 2017 Oct 13. EMBO J. 2017. PMID: 29030482 Free PMC article. - Yeast vacuoles fragment in an asymmetrical two-phase process with distinct protein requirements.
Zieger M, Mayer A. Zieger M, et al. Mol Biol Cell. 2012 Sep;23(17):3438-49. doi: 10.1091/mbc.E12-05-0347. Epub 2012 Jul 11. Mol Biol Cell. 2012. PMID: 22787281 Free PMC article.
References
- Cao H., Weller S., Orth J. D., Chen J., Huang B., Chen J. L., Stamnes M., McNiven M. A. (2005). Actin and Arf1-dependent recruitment of a cortactin-dynamin complex to the Golgi regulates post-Golgi transport. Nat. Cell Biol. 7, 483-492 - PubMed
- Cerveny K. L., Tamura Y., Zhang Z., Jensen R. E., Sesaki H. (2007). Regulation of mitochondrial fusion and division. Trends Cell Biol. 17, 563-569 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- G0601600/MRC_/Medical Research Council/United Kingdom
- BB/G011001/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- GR077544AIA/WT_/Wellcome Trust/United Kingdom
- BB/G011818/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- WT084265/WT_/Wellcome Trust/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases