Host density drives the postglacial migration of the tree parasite, Epifagus virginiana - PubMed (original) (raw)
Host density drives the postglacial migration of the tree parasite, Epifagus virginiana
Yi-Hsin Erica Tsai et al. Proc Natl Acad Sci U S A. 2010.
Abstract
To survive changes in climate, successful species shift their geographic ranges to remain in suitable habitats. For parasites and other highly specialized species, distributional changes not only are dictated by climate but can also be engineered by their hosts. The extent of host control on parasite range expansion is revealed through comparisons of host and parasite migration and demographic histories. However, understanding the codistributional history of entire forest communities is complicated by challenges in synthesizing datasets from multiple interacting species of differing datatypes. Here we integrate genetic and fossil pollen datasets from a host-parasite pair; specifically, the population structure of the parasitic plant (Epifagus virginiana) was compared with both its host (Fagus grandifolia) genetic patterns and abundance data from the paleopollen record of the last 21,000 y. Through tests of phylogeographic structure and spatial linear regression models we find, surprisingly, host range changes had little effect on the parasite's range expansion and instead host density is the main driver of parasite spread. Unlike other symbionts that have been used as proxies to track their host's movements, this parasite's migration routes are incongruent with the host and instead reflect the greater importance of host density in this community's assembly. Furthermore, these results confirm predictions of disease ecological models regarding the role of host density in the spread of pathogens. Due to host density constraints, highly specialized species may have low migration capacities and long lag times before colonization of new areas.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
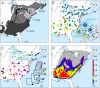
Fig. 1.
Genetic structure of Epifagus virginiana. (A) Range map with competing migration route hypotheses based on host fossil pollen (H p) or host cpDNA (H m). Regions (shaded) were defined by the distribution of host fossil pollen at 13, 9, and 6 kyBP. (B) Distribution of cpDNA haplotypes and haplotype network (Inset). Numbers are the parsimony and likelihood bootstraps and Bayesian posterior probability values. (C) Distribution of microsatellite clusters and cluster phenogram (Inset). (D) Areas of high genetic differentiation. Colors correspond to the average number of genetic breaks found at that location over 1,000 cross-validation subsamples. Color assignments were based on the 50th–90th percentiles: red, 90+%; yellow, 80–89%; green, 70–79%; blue, 60–69%; purple, 50–59%. LGM, ice margin at the last glacial maximum. Maps were drawn in Google Earth (copyright 2010 Google).
Fig. 2.
Genetic and fossil map layers of Epifagus virginiana and Fagus grandifolia. (A) Geographic range of both species. Outline shows extent of interpolated data layers. (B) Genetic distance maps for E. virginiana (EV) and F. grandifolia (F.gen). F. grandifolia genetic data are redrawn from McLachlan et al. (5). (C) Fossil pollen layers for F. grandifolia from 21 kyBP (F21) to present (F0) in 1,000-y increments. A pollen layer for 500 ybp (F0.5) is also shown. Pollen layers are redrawn from Williams et al. (6). (D) Summary pollen layers (F.age, F.avgP, and F.varP) of F. grandifolia as described in text. Warmer colors correspond with higher values in all maps. Maps were visualized using a 10-quantile color ramp.
Similar articles
- Parasitism modifies the direct effects of warming on a hemiparasite and its host.
Rafferty NE, Agnew L, Nabity PD. Rafferty NE, et al. PLoS One. 2019 Oct 30;14(10):e0224482. doi: 10.1371/journal.pone.0224482. eCollection 2019. PLoS One. 2019. PMID: 31665151 Free PMC article. - Relationships between parasite abundance and the taxonomic distance among a parasite's host species: an example with fleas parasitic on small mammals.
Krasnov BR, Shenbrot GI, Khokhlova IS, Poulin R. Krasnov BR, et al. Int J Parasitol. 2004 Oct;34(11):1289-97. doi: 10.1016/j.ijpara.2004.08.003. Int J Parasitol. 2004. PMID: 15491591 - The relationship between specialization and local abundance: the case of helminth parasites of birds.
Poulin R, Mouillot D. Poulin R, et al. Oecologia. 2004 Jul;140(2):372-8. doi: 10.1007/s00442-004-1593-4. Epub 2004 May 8. Oecologia. 2004. PMID: 15138883 - Orobanchaceae parasite-host interactions.
Mutuku JM, Cui S, Yoshida S, Shirasu K. Mutuku JM, et al. New Phytol. 2021 Apr;230(1):46-59. doi: 10.1111/nph.17083. Epub 2020 Dec 10. New Phytol. 2021. PMID: 33202061 Review.
Cited by
- Unexpectedly low rangewide population genetic structure of the imperiled eastern box turtle Terrapene c. carolina.
Kimble SJ, Rhodes OE Jr, Williams RN. Kimble SJ, et al. PLoS One. 2014 Mar 19;9(3):e92274. doi: 10.1371/journal.pone.0092274. eCollection 2014. PLoS One. 2014. PMID: 24647580 Free PMC article. - Effect of global warming on the potential distribution of a holoparasitic plant (Phelypaea tournefortii): both climate and host distribution matter.
Piwowarczyk R, Kolanowska M. Piwowarczyk R, et al. Sci Rep. 2023 Jul 3;13(1):10741. doi: 10.1038/s41598-023-37897-1. Sci Rep. 2023. PMID: 37400559 Free PMC article. - Does enemy damage vary across the range of exotic plant species? Evidence from two coastal dune plant species in eastern Australia.
Tabassum S, Leishman MR. Tabassum S, et al. Oecologia. 2018 Feb;186(2):303-309. doi: 10.1007/s00442-017-4008-z. Epub 2017 Nov 22. Oecologia. 2018. PMID: 29164370 - Plastid genomics in horticultural species: importance and applications for plant population genetics, evolution, and biotechnology.
Rogalski M, do Nascimento Vieira L, Fraga HP, Guerra MP. Rogalski M, et al. Front Plant Sci. 2015 Jul 30;6:586. doi: 10.3389/fpls.2015.00586. eCollection 2015. Front Plant Sci. 2015. PMID: 26284102 Free PMC article. Review. - The factors affecting a native obligate parasite, Cuscuta australis, in selecting an exotic weed, Humulus scandens, as its host.
Wu AP, Zhong W, Yuan JR, Qi LY, Chen FL, Liang YS, He FF, Wang YH. Wu AP, et al. Sci Rep. 2019 Jan 24;9(1):511. doi: 10.1038/s41598-018-36997-7. Sci Rep. 2019. PMID: 30679591 Free PMC article.
References
- Nieberding CM, Olivieri I. Parasites: Proxies for host genealogy and ecology? Trends Ecol Evol. 2007;22:156–165. - PubMed
- Wirth T, Meyer A, Achtman M. Deciphering host migrations and origins by means of their microbes. Mol Ecol. 2005;14:3289–3306. - PubMed
- Arneberg P, Skorping A, Grenfell B, Read AF. Host densities as determinants of abundance in parasite communities. Proc Biol Sci. 1998;265:1283–1289.
- Phillips BL, et al. Parasites and pathogens lag behind their host during periods of host range advance. Ecology. 2010;91:872–881. - PubMed
- McLachlan JS, Clark JS, Manos PS. Molecular indicators of tree migration capacity under rapid climate change. Ecology. 2005;86:2088–2098.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical