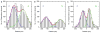In vivo imaging of labelled endogenous β-actin mRNA during nucleocytoplasmic transport - PubMed (original) (raw)
. 2010 Sep 30;467(7315):604-7.
doi: 10.1038/nature09438. Epub 2010 Sep 15.
Affiliations
- PMID: 20844488
- PMCID: PMC3005609
- DOI: 10.1038/nature09438
In vivo imaging of labelled endogenous β-actin mRNA during nucleocytoplasmic transport
David Grünwald et al. Nature. 2010.
Abstract
Export of messenger RNA occurs via nuclear pores, which are large nanomachines with diameters of roughly 120 nm that are the only link between the nucleus and cytoplasm. Hence, mRNA export occurs over distances smaller than the optical resolution of conventional light microscopes. There is extensive knowledge on the physical structure and composition of the nuclear pore complex, but transport selectivity and the dynamics of mRNA export at nuclear pores remain unknown. Here we developed a super-registration approach using fluorescence microscopy that can overcome the current limitations of co-localization by means of measuring intermolecular distances of chromatically different fluorescent molecules with nanometre precision. With this method we achieve 20-ms time-precision and at least 26-nm spatial precision, enabling the capture of highly transient interactions in living cells. Using this approach we were able to spatially resolve the kinetics of mRNA transport in mammalian cells and present a three-step model consisting of docking (80 ms), transport (5-20 ms) and release (80 ms), totalling 180 ± 10 ms. Notably, the translocation through the channel was not the rate-limiting step, mRNAs can move bi-directionally in the pore complex and not all pores are equally active.
Figures
Fig. 1
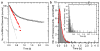
Super-Registration Precision and Detection of Nuclear mRNA. a–g) the registration precision achieved in this experiment was based on imaging nuclear pores on two cameras immediately before data acquisition (SI). Data from both cameras (a red, b green), merged image (c) after registration. A filtered merged image (d) with 21 nuclear pores, white circles outlined in red (e). f) Co-registration precision between the best aligned 6 (black bars and black line in inset), 10 (light grey) and 15 (dark grey) nuclear pores. Fit (inset), Gaussian fit to the ‘15 pore’ data set: registration= 10±1 nm, 13±1 nm FWHM. g) Distances between pores in e). Peak= 7.5 nm. h) mRNAs interacted with nuclear pores infrequently and not all interactions resulted in export of mRNAs from the nucleus (frame numbers in SI Movie 1, mRNA scanning pores, t = 40 ms. i) full length traces h) and SI Movie 1; first (blue), last (cyan). j) Intensity trace (green),tracked mRNA, background (black). k) slow export images from SI Movie 2 (frame indicated). l) fast export (SI Movie 3). m) Distances between mRNA and pore from l) colocalization precision, 26 nm total (SI). Nucleoplasmic mRNA (+) cytoplasmic mRNA (−) (SI & Fig. S5). n) Intensity mRNA signal (green) vs. background. o) mRNA positions (green boxes) and pores (red circle) overlaid on nuclear pore from l. Bars = 2 μm, ‘n/c’ = nucleus/cytoplasm, ‘max’ =maximum intensity projection, i) & o) axis pixels (= 64 nm). h–o LoG filtered (ImageJ, D. Sage).
Fig. 2
Dwell times of β-actin mRNA at the NPC. mRNA co-localized with NPCs, no. frames as milliseconds. Histogram = observed mRNAs per time bin of 20 ms. a) Fit of dwell time of cumulative trace length distribution (black circles). First bin = total number of observed traces. Fast transport events (<0.8 s) show mono-exponential decay (red circles). Dwell time = 172±3 ms, (red line, first component black line). Second time constant = 2000±120 ms is needed to fit complete data set (black line). mRNA in the nucleoplasm (grey line), dwell time = 15±1 ms (90%) and 104±6 ms (10%). Data normalized. b) Data from a) (black circles) re-plotted as trace duration histogram (black bars). Cut-off (SI &Table S3, adjacent averaging width = 5 bins). Inset = unprocessed raw data. Two-step convolution model (red line) reveals two kinetic rates, dwell times kfast = 43±1 ms and kslow = 139±10 ms. Identifying export = two observations = 40 ms. Result consistent with multistep process containing at least two rate constants, total time = 180ms.
Fig. 3
‘Binding Sites’ of mRNAs at Nuclear Pores. Distances between mRNA and POM121-tdT (zero position) bin widths = 25 nm. (−) = cytoplasmic (blue C), (+) = nucleoplasmic position (green N). Red lines are global fits, green and blue lines are fits to cytoplasmic and nucleoplasmic binding distributions. a) Histogram of all observed transport events at NPCs (b + c). b) Histogram for fast transported mRNAs (90% translocation). c) Histogram for slow mRNAs, observed for extended times at NPC.
Fig. 4
NPC Topography of mRNA Export. Results from Fig. 3B&C (hatched & open bars) plotted (a) to scale with known NPC dimensions (b). mRNA export timescale (red = kslow; orange = kfast) along NPC axis combined with single molecule data (grey bars) of Nup358, import factors and import release site. Nuclear peak position of slow transporting mRNAs located between binding sites for import factors and import release site (Fig. 4 & Table 1). Length of grey bars = FWHM of binding site distributions.
Similar articles
- An agent-based model for mRNA export through the nuclear pore complex.
Azimi M, Bulat E, Weis K, Mofrad MR. Azimi M, et al. Mol Biol Cell. 2014 Nov 5;25(22):3643-53. doi: 10.1091/mbc.E14-06-1065. Epub 2014 Sep 24. Mol Biol Cell. 2014. PMID: 25253717 Free PMC article. - To the pore and through the pore: a story of mRNA export kinetics.
Oeffinger M, Zenklusen D. Oeffinger M, et al. Biochim Biophys Acta. 2012 Jun;1819(6):494-506. doi: 10.1016/j.bbagrm.2012.02.011. Epub 2012 Feb 22. Biochim Biophys Acta. 2012. PMID: 22387213 Free PMC article. Review. - Into the basket and beyond: the journey of mRNA through the nuclear pore complex.
Ashkenazy-Titelman A, Shav-Tal Y, Kehlenbach RH. Ashkenazy-Titelman A, et al. Biochem J. 2020 Jan 17;477(1):23-44. doi: 10.1042/BCJ20190132. Biochem J. 2020. PMID: 31913454 Review. - SPEED Microscopy and Its Application in Nucleocytoplasmic Transport.
Ma J, Kelich JM, Yang W. Ma J, et al. Methods Mol Biol. 2016;1411:503-18. doi: 10.1007/978-1-4939-3530-7_31. Methods Mol Biol. 2016. PMID: 27147062 Free PMC article. - Formation of Tap/NXT1 heterodimers activates Tap-dependent nuclear mRNA export by enhancing recruitment to nuclear pore complexes.
Wiegand HL, Coburn GA, Zeng Y, Kang Y, Bogerd HP, Cullen BR. Wiegand HL, et al. Mol Cell Biol. 2002 Jan;22(1):245-56. doi: 10.1128/MCB.22.1.245-256.2002. Mol Cell Biol. 2002. PMID: 11739738 Free PMC article.
Cited by
- Tracking expression and subcellular localization of RNA and protein species using high-throughput single cell imaging flow cytometry.
Borah S, Nichols LA, Hassman LM, Kedes DH, Steitz JA. Borah S, et al. RNA. 2012 Aug;18(8):1573-9. doi: 10.1261/rna.033126.112. Epub 2012 Jun 28. RNA. 2012. PMID: 22745225 Free PMC article. - Imaging Organization of RNA Processing within the Nucleus.
Biswas J, Li W, Singer RH, Coleman RA. Biswas J, et al. Cold Spring Harb Perspect Biol. 2021 Dec 1;13(12):a039453. doi: 10.1101/cshperspect.a039453. Cold Spring Harb Perspect Biol. 2021. PMID: 34127450 Free PMC article. Review. - A jumbo problem: mapping the structure and functions of the nuclear pore complex.
Fernandez-Martinez J, Rout MP. Fernandez-Martinez J, et al. Curr Opin Cell Biol. 2012 Feb;24(1):92-9. doi: 10.1016/j.ceb.2011.12.013. Epub 2012 Feb 8. Curr Opin Cell Biol. 2012. PMID: 22321828 Free PMC article. Review. - Uncovering nuclear pore complexity with innovation.
Adams RL, Wente SR. Adams RL, et al. Cell. 2013 Mar 14;152(6):1218-21. doi: 10.1016/j.cell.2013.02.042. Cell. 2013. PMID: 23498931 Free PMC article. Review. - Pol II-directed short RNAs suppress the nuclear export of mRNA.
Komarova TV, Schwartz AM, Frolova OY, Zvereva AS, Gleba YY, Citovsky V, Dorokhov YL. Komarova TV, et al. Plant Mol Biol. 2010 Dec;74(6):591-603. doi: 10.1007/s11103-010-9700-x. Epub 2010 Oct 17. Plant Mol Biol. 2010. PMID: 20953971
References
- Stoffler D, et al. Cryo-electron tomography provides novel insights into nuclear pore architecture: Implications for nucleocytoplasmic transport. J Mol Biol. 2003;328 (1):119–130. - PubMed
- Kiseleva E, Goldberg MW, Allen TD, Akey CW. Active nuclear pore complexes in Chironomus: visualization of transporter configurations related to mRNP export. Journal of Cell Science. 1998;111:223–236. - PubMed
- Beck M, et al. Nuclear pore complex structure and dynamics revealed by cryoelectron tomography. Science. 2004;306 (5700):1387–1390. - PubMed
- Frey S, Gorlich D. A saturated FG-repeat hydrogel can reproduce the permeability properties of nuclear pore complexes. Cell. 2007;130 (3):512–523. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 GM084364-17/GM/NIGMS NIH HHS/United States
- R01 GM086217-04/GM/NIGMS NIH HHS/United States
- R01 GM084364-18/GM/NIGMS NIH HHS/United States
- R01 GM086217/GM/NIGMS NIH HHS/United States
- R01 GM084364-16/GM/NIGMS NIH HHS/United States
- EB2060/EB/NIBIB NIH HHS/United States
- R01 GM084364/GM/NIGMS NIH HHS/United States
- R01 GM084364-15A1/GM/NIGMS NIH HHS/United States
- R01 EB002060-20/EB/NIBIB NIH HHS/United States
- GM86217/GM/NIGMS NIH HHS/United States
- R01 EB002060/EB/NIBIB NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials