Chromatin landscape dictates HSF binding to target DNA elements - PubMed (original) (raw)
Chromatin landscape dictates HSF binding to target DNA elements
Michael J Guertin et al. PLoS Genet. 2010.
Abstract
Sequence-specific transcription factors (TFs) are critical for specifying patterns and levels of gene expression, but target DNA elements are not sufficient to specify TF binding in vivo. In eukaryotes, the binding of a TF is in competition with a constellation of other proteins, including histones, which package DNA into nucleosomes. We used the ChIP-seq assay to examine the genome-wide distribution of Drosophila Heat Shock Factor (HSF), a TF whose binding activity is mediated by heat shock-induced trimerization. HSF binds to 464 sites after heat shock, the vast majority of which contain HSF Sequence-binding Elements (HSEs). HSF-bound sequence motifs represent only a small fraction of the total HSEs present in the genome. ModENCODE ChIP-chip datasets, generated during non-heat shock conditions, were used to show that inducibly bound HSE motifs are associated with histone acetylation, H3K4 trimethylation, RNA Polymerase II, and coactivators, compared to HSE motifs that remain HSF-free. Furthermore, directly changing the chromatin landscape, from an inactive to an active state, permits inducible HSF binding. There is a strong correlation of bound HSEs to active chromatin marks present prior to induced HSF binding, indicating that an HSE's residence in "active" chromatin is a primary determinant of whether HSF can bind following heat shock.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
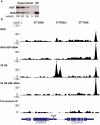
Figure 1. HSF depletion filters false positive peaks.
(A) Densitometry of the loading control (TFIIS) confirmed that the intensity of each band was proportional to the number of cells loaded. The HSF-KD HSF band is 1.6 times the intensity of the most dilute HSF band of the standard curve, indicating a 40-fold depletion of HSF. (B) The UCSC Genome Browser is used to show a locus that contains two legitimate HS inducible/HSF-RNAi-sensitive binding sites (represented by asterisks) and a false-positive peak that is neither inducible nor sensitive to HSF depletion (represented by “×”). The y-axis scale is linear (from 2 to 110) and normalized for each experiment (shifted tags/10 bp/10 million sequences in the library). Mock IP with the pre-inoculated animal serum served as a background dataset (Pre-immune IP).
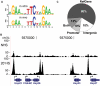
Figure 2. Characterization of HSF binding sites.
(A) The PSWM derived from in vitro band shift assays (top) and this study (bottom) are compared. Sequence logos were generated using WebLogo . (B) The 67B locus harbors known heat shock protein (hsp) genes. The y-axis scale is linear (from 2 to 180) and directly comparable for each condition (shifted tags/10 bp/10 million sequences in the library). HSF binding sites, detected by our peak calling criteria (asterisks), increased in signal intensity or appeared de novo as cells were shifted from NHS to HS. (C) HSF binding sites are found within the body of RefGenes (72%: 316 sites), in the 500 bases upstream of TSS (22%: 97 sites), and within intergenic regions (18%: 81). A precise genomic sequence can be both within a gene and within the promoter of an upstream gene; 52 binding sites (the 12% slice) fall in this category.
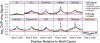
Figure 3. Bound HSE motifs contain marks of active chromatin prior to HSF binding.
The average factor or histone modification occupancy was assigned in 100 base windows (step size of 50) around HSF-free HSE motifs (red) and HSF-bound HSE motifs (green). HSF-bound motifs are categorized by annotation class: motifs within promoters (magenta), RefGene bodies (blue), and intergenic regions (black). Canonical active chromatin marks are enriched at HSF-bound motifs (purple). H3K27me3 is depleted at HSF-bound motifs (orange).
Figure 4. Bound HSE motifs are statistically associated with marks of active chromatin, compared to HSF-free motifs.
For each factor shown, the Fisher exact test was used to determine the statistically significant association of HSF-bound motifs (left bar in each panel) with each modENCODE factor or histone modification, compared to HSF-free motifs (right bar). The yellow fraction of the bar chart represents HSF binding sites that are within regions of significant enrichment, while blue depicts all non-enriched sites.
Figure 5. Active chromatin marks are clustered at HSE motifs.
K-means clustering analysis, specifying five clusters, reveals that the histone modifications tend to occur together at HSF-bound motifs. Each motif corresponds to an individual row. Columns represent the average microarray intensity of all the probes in a 400 base window centered on the motif for a given factor or histone modification. Cluster and Treeview were used to generate and visualize the clustering data , .
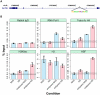
Figure 6. Changing the chromatin landscape converts an HSF-free motif to an HSF-bound site.
(A) The ecdysone inducible gene Eip75B harbors an HSF motif that conforms to the consensus with a p-value of 1.2×10−7. (B) The blue bars represent the changes in factor and histone modification occupancy after ecdysone is added to the cells. The pink bars indicate the changes in occupancy after HS treatment in cells that were pre-treated with ecdysone. Precipitation with Rabbit IgG controls for non-specific pull-down at this site for each condition (first sub-panel) and dashed and solid lines indicate the range of background intensities for non-specific background pull-down by each antibody (see Materials and Methods) and provides an estimated threshold for assessing enrichment over background.
Figure 7. Induced mRNA accumulation after a 20′ HS shows that promoter-bound HSF has varying induction effects.
Oligo dT-reverse transcribed RNA was subjected to real-time qPCR with the primers illustrated in Figure S15 (sequences available within Dataset S6), during NHS and 20′ HS conditions. All mRNA levels were normalized to RpL32 and are represented as HS mRNA levels divided by NHS levels. Three independent biological replicates and two technical replicates for each biological sample were performed.
Similar articles
- Accurate prediction of inducible transcription factor binding intensities in vivo.
Guertin MJ, Martins AL, Siepel A, Lis JT. Guertin MJ, et al. PLoS Genet. 2012;8(3):e1002610. doi: 10.1371/journal.pgen.1002610. Epub 2012 Mar 29. PLoS Genet. 2012. PMID: 22479205 Free PMC article. - Cooperative binding of heat shock transcription factor to the Hsp70 promoter in vivo and in vitro.
Amin J, Fernandez M, Ananthan J, Lis JT, Voellmy R. Amin J, et al. J Biol Chem. 1994 Feb 18;269(7):4804-11. J Biol Chem. 1994. PMID: 8106450 - HSF access to heat shock elements in vivo depends critically on promoter architecture defined by GAGA factor, TFIID, and RNA polymerase II binding sites.
Shopland LS, Hirayoshi K, Fernandes M, Lis JT. Shopland LS, et al. Genes Dev. 1995 Nov 15;9(22):2756-69. doi: 10.1101/gad.9.22.2756. Genes Dev. 1995. PMID: 7590251 - Whole-genome analysis reveals that active heat shock factor binding sites are mostly associated with non-heat shock genes in Drosophila melanogaster.
Gonsalves SE, Moses AM, Razak Z, Robert F, Westwood JT. Gonsalves SE, et al. PLoS One. 2011 Jan 14;6(1):e15934. doi: 10.1371/journal.pone.0015934. PLoS One. 2011. PMID: 21264254 Free PMC article. - Drosophila heat shock system as a general model to investigate transcriptional regulation.
Guertin MJ, Petesch SJ, Zobeck KL, Min IM, Lis JT. Guertin MJ, et al. Cold Spring Harb Symp Quant Biol. 2010;75:1-9. doi: 10.1101/sqb.2010.75.039. Epub 2011 Apr 5. Cold Spring Harb Symp Quant Biol. 2010. PMID: 21467139 Free PMC article. Review.
Cited by
- Mechanistic Insights Into the Interaction Between Transcription Factors and Epigenetic Modifications and the Contribution to the Development of Obesity.
Huang Q, Ma C, Chen L, Luo D, Chen R, Liang F. Huang Q, et al. Front Endocrinol (Lausanne). 2018 Jul 6;9:370. doi: 10.3389/fendo.2018.00370. eCollection 2018. Front Endocrinol (Lausanne). 2018. PMID: 30034368 Free PMC article. Review. - High levels of nuclear heat-shock factor 1 (HSF1) are associated with poor prognosis in breast cancer.
Santagata S, Hu R, Lin NU, Mendillo ML, Collins LC, Hankinson SE, Schnitt SJ, Whitesell L, Tamimi RM, Lindquist S, Ince TA. Santagata S, et al. Proc Natl Acad Sci U S A. 2011 Nov 8;108(45):18378-83. doi: 10.1073/pnas.1115031108. Epub 2011 Oct 31. Proc Natl Acad Sci U S A. 2011. PMID: 22042860 Free PMC article. - Nuclear receptor signaling via NHR-49/MDT-15 regulates stress resilience and proteostasis in response to reproductive and metabolic cues.
Sala AJ, Grant RA, Imran G, Morton C, Brielmann RM, Gorgoń S, Watts J, Bott LC, Morimoto RI. Sala AJ, et al. Genes Dev. 2024 Jun 25;38(9-10):380-392. doi: 10.1101/gad.351829.124. Genes Dev. 2024. PMID: 38816072 Free PMC article. - Impact of heat shock transcription factor 1 on global gene expression profiles in cells which induce either cytoprotective or pro-apoptotic response following hyperthermia.
Kus-Liśkiewicz M, Polańska J, Korfanty J, Olbryt M, Vydra N, Toma A, Widłak W. Kus-Liśkiewicz M, et al. BMC Genomics. 2013 Jul 8;14:456. doi: 10.1186/1471-2164-14-456. BMC Genomics. 2013. PMID: 23834426 Free PMC article. - Primordial super-enhancers: heat shock-induced chromatin organization in yeast.
Kainth AS, Chowdhary S, Pincus D, Gross DS. Kainth AS, et al. Trends Cell Biol. 2021 Oct;31(10):801-813. doi: 10.1016/j.tcb.2021.04.004. Epub 2021 May 14. Trends Cell Biol. 2021. PMID: 34001402 Free PMC article. Review.
References
- Wei CL, Wu Q, Vega VB, Chiu KP, Ng P, et al. A global map of p53 transcription-factor binding sites in the human genome. Cell. 2006;124:207–219. - PubMed
- Yang A, Zhu Z, Kapranov P, McKeon F, Church GM, et al. Relationships between p63 binding, DNA sequence, transcription activity, and biological function in human cells. Mol Cell. 2006;24:593–602. - PubMed
- Biggin MD, McGinnis W. Regulation of segmentation and segmental identity by Drosophila homeoproteins: The role of DNA binding in functional activity and specificity. Development. 1997;124:4425–4433. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- T32 GM007617/GM/NIGMS NIH HHS/United States
- GM25232/GM/NIGMS NIH HHS/United States
- R37 GM025232/GM/NIGMS NIH HHS/United States
- T32-GM007617/GM/NIGMS NIH HHS/United States
- R01 GM025232/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous