Targeting DNA methylation for epigenetic therapy - PubMed (original) (raw)
Review
Targeting DNA methylation for epigenetic therapy
Xiaojing Yang et al. Trends Pharmacol Sci. 2010 Nov.
Abstract
Patterns of DNA methylation are established during embryonic development and faithfully copied through somatic cell divisions. Based on current understanding of DNA methylation and other interrelated epigenetic modifications, a comprehensive view of the 'epigenetic landscape' and cancer epigenome is evolving. The cancer methylome is highly disrupted, making DNA methylation an excellent target for anticancer therapies. During the last few decades, an increasing number of drugs targeting DNA methylation have been developed to increase efficacy and stability and to decrease toxicity. The earliest and the most successful epigenetic drug to date, 5-Azacytidine, is currently recommended as the first-line treatment of high-risk myelodysplastic syndromes (MDS). Encouraging results from clinical trials have prompted further efforts to elucidate epigenetic alterations in cancer, and to subsequently develop new epigenetic therapies. This review delineates the latest cancer epigenetic models, the recent discovery of hypomethylation agents as well as their application in the clinic.
Copyright © 2010 Elsevier Ltd. All rights reserved.
Figures
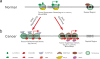
Figure 1. Epigenetic Regulation in Normal and Cancer Cells
Shown are schematic promoters. Arrows represent transcription start site (TSS); filled circles represent methylated CpG dinucleotide and empty circles represent unmethylated CpG dinucleotide. In normal cells (a), genes such as MLH1, CDKN2A are generally unmethylated and packaged with active modified histone proteins (e.g. H3K4me3) as well as histone variants (e.g. H2A.Z). These epigenetic modifications constitute an “open” chromatin structure which, with nucleosome depleted region (NDR), favoring transcription. In other genomic regions, such as in the repetitive elements, the CpG sites are methylated and thereby maintain a closed chromatin structure. In cancer cells (b), epigenetic modifications are disrupted. Besides cancer specific hypomethylation (e.g. in repetitive sequence), there are two interrelated epigenetic mechanisms to repress gene expression. Some genes (e.g.FBXO32) could be recognized by polycomb proteins, such as EZH2, which catalyses H3K27 methylation, and are consequently repressed. By contrast, CpG sites within gene promoters could undergo _de nov_o methylation by DNMT3A/B, which are compartmentalized to these regions to complete the methylation process. The methylated CpG sites attract methyl-binding proteins such as MBD, which is coupled with HDAC proteins to remove histone acetylation as well as histone methyltransferase to methylate H3K9. Associated with all repressive factors shown, nucleosomes cover the promoter region, generating a tightly closed chromatin status to shut down gene expression.
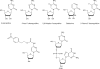
Figure 2. Chemical Structure of DNMT inhibitors
A. nucleoside analogues B. non-nucleoside analogues.
Figure 2. Chemical Structure of DNMT inhibitors
A. nucleoside analogues B. non-nucleoside analogues.
Figure 3. Metabolism pathway and action mechanism of 5-Azanucleosides
A. Human Concentrated Nucleoside Transporters 1 (hCNT1) facilitates the entry of 5-Aza-CR and 5-Aza-CdR into cells where they are phosphorylated by uridine-cytidine kinase and deoxycytidine kinase respectively. 5-Aza-CMP and 5-Aza-dCMP are subsequently phosphorylated into their active triphosphate forms. 5 Aza-dCR can be incorporated solely into DNA, whereas 5-Aza-CR can be incorporated into RNA as well as DNA following the reduction of its 5-Aza-CDP form to the 5-Aza-dCDP form. Incorporation of 5-Azanucleosides into DNA induces hypomethylation of the daughter DNA strands whereas incorporation of 5-Aza-CR into RNA disrupts crucial cellular process such as protein translation and causes ribosomal disassembly. 5-Azanucleosides, however, are also very unstable and can be deaminated to inactive uridine. B. Schematic working model of 5-Azanucleosides. During DNA replication, 5-Azanucleosides are incorporated into DNA and trap DNMTs, which are subsequently targeted for proteosomal degradation. DNA containing azanucleosides are hemimethylated after the first round of DNA replication and become fully demethylated after several rounds of replication. Using hypomethylation agents, the silenced epigenetic modifications could be switched to an active status (eg.H3K4me3 and AcH3)
Similar articles
- Will next-generation agents deliver on the promise of epigenetic hypomethylation therapy?
Lowder JN, Taverna P, Issa JP. Lowder JN, et al. Epigenomics. 2015 Oct;7(7):1083-8. doi: 10.2217/epi.15.66. Epub 2015 Nov 6. Epigenomics. 2015. PMID: 26541345 No abstract available. - Targeting DNA methylation.
Issa JP, Kantarjian HM. Issa JP, et al. Clin Cancer Res. 2009 Jun 15;15(12):3938-46. doi: 10.1158/1078-0432.CCR-08-2783. Epub 2009 Jun 9. Clin Cancer Res. 2009. PMID: 19509174 Free PMC article. - [Epigenetic therapy in myelodysplastic syndromes (MDS). Treatment with DNA methyltransferase inhibitors].
Daskalakis M, Blagitko-Dorfs N, Hackanson B, Lübbert M. Daskalakis M, et al. Pharm Unserer Zeit. 2010 May;39(3):217-27. doi: 10.1002/pauz.201000369. Pharm Unserer Zeit. 2010. PMID: 20425776 Review. German. No abstract available. - DNA hypermethylation as a chemotherapy target.
Ren J, Singh BN, Huang Q, Li Z, Gao Y, Mishra P, Hwa YL, Li J, Dowdy SC, Jiang SW. Ren J, et al. Cell Signal. 2011 Jul;23(7):1082-93. doi: 10.1016/j.cellsig.2011.02.003. Epub 2011 Feb 21. Cell Signal. 2011. PMID: 21345368 Review. - Epigenetic drug discovery: targeting DNA methyltransferases.
Foulks JM, Parnell KM, Nix RN, Chau S, Swierczek K, Saunders M, Wright K, Hendrickson TF, Ho KK, McCullar MV, Kanner SB. Foulks JM, et al. J Biomol Screen. 2012 Jan;17(1):2-17. doi: 10.1177/1087057111421212. Epub 2011 Sep 30. J Biomol Screen. 2012. PMID: 21965114 Review.
Cited by
- Direct but no transgenerational effects of decitabine and vorinostat on male fertility.
Kläver R, Sánchez V, Damm OS, Redmann K, Lahrmann E, Sandhowe-Klaverkamp R, Rohde C, Wistuba J, Ehmcke J, Schlatt S, Gromoll J. Kläver R, et al. PLoS One. 2015 Feb 18;10(2):e0117839. doi: 10.1371/journal.pone.0117839. eCollection 2015. PLoS One. 2015. PMID: 25692788 Free PMC article. - Isoform switching and exon skipping induced by the DNA methylation inhibitor 5-Aza-2'-deoxycytidine.
Ding XL, Yang X, Liang G, Wang K. Ding XL, et al. Sci Rep. 2016 Apr 19;6:24545. doi: 10.1038/srep24545. Sci Rep. 2016. PMID: 27090213 Free PMC article. - Identification of SDG gene family members and exploration of flowering related genes in different cultivars of chrysanthemums and their wild ancestors.
Han T, Khan MA, Wang Y, Tan W, Li C, Ai P, Zhao W, Li Z, Wang Z. Han T, et al. BMC Plant Biol. 2024 Aug 29;24(1):813. doi: 10.1186/s12870-024-05465-y. BMC Plant Biol. 2024. PMID: 39210253 Free PMC article. - Epigenetic Approaches to the Treatment of Dental Pulp Inflammation and Repair: Opportunities and Obstacles.
Kearney M, Cooper PR, Smith AJ, Duncan HF. Kearney M, et al. Front Genet. 2018 Aug 7;9:311. doi: 10.3389/fgene.2018.00311. eCollection 2018. Front Genet. 2018. PMID: 30131827 Free PMC article. Review. - The Role of Cell-Free DNA in Cancer Treatment Decision Making.
Telekes A, Horváth A. Telekes A, et al. Cancers (Basel). 2022 Dec 12;14(24):6115. doi: 10.3390/cancers14246115. Cancers (Basel). 2022. PMID: 36551600 Free PMC article. Review.
References
- Suzuki MM, Bird A. DNA methylation landscapes: provocative insights from epigenomics. Nat Rev Genet. 2008;9:465–476. - PubMed
- Wang Y, Leung FC. An evaluation of new criteria for CpG islands in the human genome as gene markers. Bioinformatics. 2004;20:1170–1177. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous