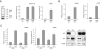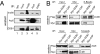INO80 chromatin remodeling complex promotes the removal of UV lesions by the nucleotide excision repair pathway - PubMed (original) (raw)
INO80 chromatin remodeling complex promotes the removal of UV lesions by the nucleotide excision repair pathway
Yingjun Jiang et al. Proc Natl Acad Sci U S A. 2010.
Abstract
The creation of accessible DNA in the context of chromatin is a key step in many DNA functions. To reveal how ATP-dependent chromatin remodeling activities impact DNA repair, we constructed mammalian genetic models for the INO80 chromatin remodeling complex and investigated the impact of loss of INO80 function on the repair of UV-induced photo lesions. We showed that deletion of two core components of the INO80 complex, INO80 and ARP5, significantly hampered cellular removal of UV-induced photo lesions but had no significant impact on the transcription of nucleotide excision repair (NER) factors. Loss of INO80 abolished the assembly of NER factors, suggesting that prior chromatin relaxation is important for the NER incision process. Ino80 and Arp5 are enriched to UV-damaged DNA in an NER-incision-independent fashion, suggesting that recruitment of the remodeling activity likely takes place during the initial stage of damage recognition. These results demonstrate a critical role of INO80 in creating DNA accessibility for the NER pathway and provide direct evidence that repair of UV lesions and perhaps most bulky adduct lesions requires chromatin reconfiguration.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Fig. 1.
Conditional inactivation of the INO80 locus. (A) Genotype of the INO80Flox/− conditional mutant with one allele carrying floxed exons 2–4 and the other allele with an in-frame insertion of the Neo-coding sequence. AdCre infection results in the inactivation of the conditional allele. (B) Depletion of the Ino80 protein from the INO80Flox/− conditional mutant. Immunoblotting of Ino80 was performed with an affinity-purified polyclonal antibody and cell extracts were prepared 3 d after AdCre infection. β-Actin was used as a loading control. 4H12 and 2D15 are two independently derived conditional mutants. (C) Depletion of the Arp5 protein from the ARP5Flox/− conditional mutant. Immunoblotting was performed with an affinity-purified polyclonal antibody and cell extracts were prepared 3 d after AdCre infection. β-Actin was used as a loading control. 7B1 and 5E9 are two independently derived conditional mutants. (D) MTT assays on INO80 and ARP5 conditional mutants 5 d after AdCre treatment. Error bars represent SD from three independent experiments. (E) [3H]Thymidine incorporation of INO80+/+ and INO80−/− cells 4 d after AdCre infection. Error bars represent SD from three independent samples.
Fig. 2.
Defective repair of UV photo lesions in cells lacking INO80 and transcriptional impact of INO80 depletion. (A) Slot blotting of CPD (Left) in genomic DNA isolated from INO80 WT and INO80 mutant cells (4H12 and 2D15) irradiated with 8 J/m2 of UV (254 nm) and harvested at 0, 6, and 24 h. Quantifications of the relative CPD levels were arrived at by normalizing the chemiluminescent signal against the total DNA signal obtained by Southern hybridization with a total genomic DNA probe (Right). (B) Slot blotting of 6-4PP (Left) in genomic DNA isolated from INO80 WT and INO80 mutant cells (4H12 and 2D15) irradiated with 8 J/m2 of UV (254 nm) and harvested at 0, 1, 3, and 6 h. Quantifications of the relative 6-4PP levels were similarly performed as in A. (C) Immunoblotting of NER proteins in INO80+/+ and INO80−/− cells. (D) In vitro NER activity of INO80 and ARP5 mutants. Nuclear extracts were prepared from wild-type (INO80+/+), heterozygous (P1G8, INO80Flox/+), and two conditional INO80Flox/− cells (4H12 and 2D15) treated (+) and untreated (−) with AdCre and used in the NER synthesis assay. (E) Repair synthesis assay with nuclear extracts prepared from WT (ARP5+/+) and two conditional ARP5Flox/− mutants (7B1 and 5E9) treated (+) or untreated (−) with AdCre. (Upper) DNA substrates from each sample were recovered after the repair synthesis assay and resolved on an agarose gel. (Lower) Autoradiography generated by PhosphorImager (Molecular Dynamics). Relative NER activity of each sample (Bottom) was arrived at by normalizing the [32P]dCTP incorporation in damaged plasmids against each internal control.
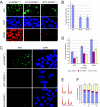
Fig. 3.
XPC and XPA foci formation in INO80-deficient cells. (A) Formation of UV-induced XPC foci in INO80 mutant cells 30 min after UV exposure (2,000 J/m2). (B) Quantification of XPC foci-positive cells in WT and INO80−/− cells. (C) Formation of UV-induced XPA foci in INO80 mutant cells. (D) Quantification of XPA foci-positive cells in WT and INO80−/− cells. UV-induced foci were generated by localized irradiation with 5-μm (XPC) and 2-μm (XPA) Isopore filters (Millipore). (E) WT (INO80+/+) and 2D15 (INO80−/−) cells were mock-treated or treated (+ UV) with 50 J/m2. Cell-cycle profiles were measured 1 h after treatment by flow cytometry. Representative data from two experiments are shown. (F) Quantification of E.
Fig. 4.
UV damage-specific recruitment of the INO80 complex detected by ChIP analysis. (A) Plasmid DNA (pOriP) was UV-irradiated (UV+) or mock-treated (UV−) to induced UV lesions as detected by slot blotting with an anti-64-PP antibody (Left) and electroporated into HCT116 cells. ChIP analyses were carried out 0.5 h after transfection with anti-histone H3 (Middle) and ERCC1 (Right) antibodies, as indicated. Percentages of relative enrichment of each protein to UV-damaged DNA or mock-treated DNA were arrived at by normalizing comparative concentrations of each sample with that of its input lysate. (B) Recruitment of INO80 and ARP5 proteins to UV-treated and mock-treated plasmid DNA. (C) UV damage-dependent recruitment of INO80 in XPA/XP2OS (XPA−) (Left) and XPC/XP4PA (XPC−) (Right) mutants and their isogenic, complemented controls (XPA+ and XPC+, respectively). Error bars representing SD were derived from three or more experiments with triplicate quantitative PCR reactions. The P values were derived from paired t tests.
Fig. 5.
Interaction between the Ino80 complex and NER factors. (A) Immunoprecipitation of Ino80 and Arp5 from untreated HCT116 cell extracts. Endogenous proteins precipitated with the indicated antibodies were immunoblotted with antibodies against Ino80, Arp5, and DDB1. (B) Reciprocal IP between DDB1 and Ino80 complex components. Myc-DDB1 (125 kDa) or the negative control Myc-CtIP (125 kDa) was coexpressed with SFB-tagged Ino80 (Upper) or SFB-tagged Arp5 (Lower). (Upper, Middle) Cell lysates were immunoprecipitated with an anti-Myc antibody followed by immunoblotting with anti-Myc and anti-Flag antibodies. (Upper, Right) Cell lysates were subjected to streptavidin-bead (S-beads) pull-down assays to recover SFB-Ino80 and its associated proteins, followed by immunoblotting with an anti-Myc antibody. Similarly, detection of Myc-DDB1–Arp5 interaction was performed (Right). Coimmunoprecipitations and pull-down assays were performed in the presence of ethidium bromide (50 μg/mL).
Similar articles
- The Ino80 chromatin-remodeling complex restores chromatin structure during UV DNA damage repair.
Sarkar S, Kiely R, McHugh PJ. Sarkar S, et al. J Cell Biol. 2010 Dec 13;191(6):1061-8. doi: 10.1083/jcb.201006178. Epub 2010 Dec 6. J Cell Biol. 2010. PMID: 21135142 Free PMC article. - INO80 and gamma-H2AX interaction links ATP-dependent chromatin remodeling to DNA damage repair.
Morrison AJ, Highland J, Krogan NJ, Arbel-Eden A, Greenblatt JF, Haber JE, Shen X. Morrison AJ, et al. Cell. 2004 Dec 17;119(6):767-75. doi: 10.1016/j.cell.2004.11.037. Cell. 2004. PMID: 15607974 - Recruitment of the INO80 complex by H2A phosphorylation links ATP-dependent chromatin remodeling with DNA double-strand break repair.
van Attikum H, Fritsch O, Hohn B, Gasser SM. van Attikum H, et al. Cell. 2004 Dec 17;119(6):777-88. doi: 10.1016/j.cell.2004.11.033. Cell. 2004. PMID: 15607975 - Assays for chromatin remodeling during nucleotide excision repair in Saccharomyces cerevisiae.
Zhang L, Jones K, Smerdon MJ, Gong F. Zhang L, et al. Methods. 2009 May;48(1):19-22. doi: 10.1016/j.ymeth.2009.03.017. Epub 2009 Mar 29. Methods. 2009. PMID: 19336254 Free PMC article. Review. - Nucleotide excision repair in chromatin: damage removal at the drop of a HAT.
Reed SH. Reed SH. DNA Repair (Amst). 2011 Jul 15;10(7):734-42. doi: 10.1016/j.dnarep.2011.04.029. Epub 2011 May 20. DNA Repair (Amst). 2011. PMID: 21600858 Review.
Cited by
- INO80 and SWR1 complexes: the non-identical twins of chromatin remodelling.
Willhoft O, Wigley DB. Willhoft O, et al. Curr Opin Struct Biol. 2020 Apr;61:50-58. doi: 10.1016/j.sbi.2019.09.002. Epub 2019 Dec 12. Curr Opin Struct Biol. 2020. PMID: 31838293 Free PMC article. Review. - Fluorescently-labelled CPD and 6-4PP photolyases: new tools for live-cell DNA damage quantification and laser-assisted repair.
Steurer B, Turkyilmaz Y, van Toorn M, van Leeuwen W, Escudero-Ferruz P, Marteijn JA. Steurer B, et al. Nucleic Acids Res. 2019 Apr 23;47(7):3536-3549. doi: 10.1093/nar/gkz035. Nucleic Acids Res. 2019. PMID: 30698791 Free PMC article. - Epigenetic regulation of genomic integrity.
Deem AK, Li X, Tyler JK. Deem AK, et al. Chromosoma. 2012 Apr;121(2):131-51. doi: 10.1007/s00412-011-0358-1. Epub 2012 Jan 17. Chromosoma. 2012. PMID: 22249206 Free PMC article. Review. - Chromatin remodeler CHD1 promotes XPC-to-TFIIH handover of nucleosomal UV lesions in nucleotide excision repair.
Rüthemann P, Balbo Pogliano C, Codilupi T, Garajovà Z, Naegeli H. Rüthemann P, et al. EMBO J. 2017 Nov 15;36(22):3372-3386. doi: 10.15252/embj.201695742. Epub 2017 Oct 10. EMBO J. 2017. PMID: 29018037 Free PMC article.
References
- Lusser A, Kadonaga JT. Chromatin remodeling by ATP-dependent molecular machines. Bioessays. 2003;25:1192–1200. - PubMed
- Owen-Hughes T, et al. Analysis of nucleosome disruption by ATP-driven chromatin remodeling complexes. Methods Mol Biol. 1999;119:319–331. - PubMed
- Peterson CL, Côté J. Cellular machineries for chromosomal DNA repair. Genes Dev. 2004;18:602–616. - PubMed
- Ebbert R, Birkmann A, Schüller H-J. The product of the SNF2/SWI2 paralogue INO80 of Saccharomyces cerevisiae required for efficient expression of various yeast structural genes is part of a high-molecular-weight protein complex. Mol Microbiol. 1999;32:741–751. - PubMed
- Shen X, Mizuguchi G, Hamiche A, Wu C. A chromatin remodelling complex involved in transcription and DNA processing. Nature. 2000;406:541–544. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- CA97175/CA/NCI NIH HHS/United States
- P01 CA097175/CA/NCI NIH HHS/United States
- R01 GM093104/GM/NIGMS NIH HHS/United States
- R01 CA079648/CA/NCI NIH HHS/United States
- R01 CA127945-04/CA/NCI NIH HHS/United States
- CA 127945/CA/NCI NIH HHS/United States
- R01 CA127945/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases