Social network sensors for early detection of contagious outbreaks - PubMed (original) (raw)
Social network sensors for early detection of contagious outbreaks
Nicholas A Christakis et al. PLoS One. 2010.
Abstract
Current methods for the detection of contagious outbreaks give contemporaneous information about the course of an epidemic at best. It is known that individuals near the center of a social network are likely to be infected sooner during the course of an outbreak, on average, than those at the periphery. Unfortunately, mapping a whole network to identify central individuals who might be monitored for infection is typically very difficult. We propose an alternative strategy that does not require ascertainment of global network structure, namely, simply monitoring the friends of randomly selected individuals. Such individuals are known to be more central. To evaluate whether such a friend group could indeed provide early detection, we studied a flu outbreak at Harvard College in late 2009. We followed 744 students who were either members of a group of randomly chosen individuals or a group of their friends. Based on clinical diagnoses, the progression of the epidemic in the friend group occurred 13.9 days (95% C.I. 9.9-16.6) in advance of the randomly chosen group (i.e., the population as a whole). The friend group also showed a significant lead time (p<0.05) on day 16 of the epidemic, a full 46 days before the peak in daily incidence in the population as a whole. This sensor method could provide significant additional time to react to epidemics in small or large populations under surveillance. The amount of lead time will depend on features of the outbreak and the network at hand. The method could in principle be generalized to other biological, psychological, informational, or behavioral contagions that spread in networks.
Conflict of interest statement
Competing Interests: The authors have an equity stake in a company, MedNetworks, that is licensed by Harvard and UCSD to apply some of the ideas embodied in their work.
Figures
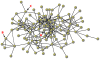
Figure 1. Network Illustrating Structural Parameters.
This real network of 105 students shows variation in structural attributes and topological position. Each circle represents a person and each line represents a friendship tie. Nodes A and B have different “degree,” a measure that indicates the number of ties. Nodes with higher degree also tend to exhibit higher “centrality” (node A with six friends is more central than B and C who both only have four friends). If contagions infect people at random at the beginning of an epidemic, central individuals are likely to be infected sooner because they lie a shorter number of steps (on average) from all other individuals in the network. Finally, although nodes B and C have the same degree, they differ in “transitivity” (the probability that any two of one's friends are friends with each other). Node B exhibits high transitivity with many friends that know one another. In contrast, node C's friends are not connected to one another and therefore they offer more independent possibilities for becoming infected earlier in the epidemic.
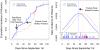
Figure 2. Theoretical expectations of differences in contagion between central individuals and the population as a whole.
A contagious process passes through two phases, one in which the number of infected individuals exponentially increases as the contagion spreads, and one in which incidence exponentially decreases as susceptible individuals become increasingly scarce. These dynamics can be modeled by a logistic function. Central individuals lie on more paths in a network compared to the average person in a population and are therefore more likely to be infected early by a contagion that randomly infects some individuals and then spreads from person to person within the network. This shifts the S-shaped logistic cumulative incidence function forward in time for central individuals compared to the population as a whole (left panel). It also shifts the peak infection rate forward (right panel).
Figure 3. Empirical differences in flu contagion between “friend” group and randomly chosen individuals.
We compared two groups, one composed of individuals randomly selected from our population, and one composed of individuals who were nominated as a friend by members of the random group. The friend group was observed to have significantly higher measured in-degree and betweenness centrality than the random group (see Supporting Information Text S1). In the left panel, a nonparametric maximum likelihood estimate (NPMLE) of cumulative flu incidence (based on diagnoses by medical staff) shows that individuals in the friend group tended to get the flu earlier than individuals in the random group. Moreover, predicted daily incidence from a nonlinear least squares fit of the data to a logistic distribution function suggests that the peak incidence of flu is shifted forward in time for the friends group by 13.9 days (right panel). A significant (p<0.05) lead time for the friend group was first detected with data available up to Day 16. Raw data for daily flu cases in the friend group (blue) and random group (red) is shown in the inset box (right panel).
Figure 4. Progression of flu contagion in the friendship network over time.
Each frame shows the largest component of the network (714 people) for a specific date, with each line representing a friendship nomination and each node representing a person. Infected individuals are colored red, friends of infected individuals are colored yellow, and node size is proportional to the number of friends infected. All available information regarding infections is used here. Frames for all 122 days of the study are available in a movie of the epidemic posted in the Supporting Information (Video S1).
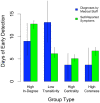
Figure 5. Estimated days of advance detection of a flu outbreak when following specific groups.
Here, degree, transitivity, centrality, and coreness are computed based on the mapping of the network. The high in-degree group is composed of individuals who have a higher-than-average number of other people in the network who name them as a friend. The low transitivity group is composed of individuals with below-average probability that any two of their friends are friends with one another. The high centrality group is composed of individuals with a higher-than-average betweenness, which is the number of shortest paths connecting all individuals in a network that pass through a given person. The high coreness group is composed of individuals with a higher-than-average coreness, which is the number of friends a person has once all individuals with fewer friends have been eliminated from the network. Analyses were conducted separately for data based on flu diagnoses by medical staff (blue bars) and data based on self-reported flu symptoms (green bars). Estimates and 95% confidence intervals are based on a nonlinear least squares fit of the flu data to a logistic distribution function (see Supporting Information Text S1). The results show that flu outbreaks occur up to two weeks earlier in each of these groups.
Similar articles
- Using friends as sensors to detect global-scale contagious outbreaks.
Garcia-Herranz M, Moro E, Cebrian M, Christakis NA, Fowler JH. Garcia-Herranz M, et al. PLoS One. 2014 Apr 9;9(4):e92413. doi: 10.1371/journal.pone.0092413. eCollection 2014. PLoS One. 2014. PMID: 24718030 Free PMC article. - Efficient detection of contagious outbreaks in massive metropolitan encounter networks.
Sun L, Axhausen KW, Lee DH, Cebrian M. Sun L, et al. Sci Rep. 2014 Jun 6;4:5099. doi: 10.1038/srep05099. Sci Rep. 2014. PMID: 24903017 Free PMC article. - Dueling biological and social contagions.
Fu F, Christakis NA, Fowler JH. Fu F, et al. Sci Rep. 2017 Mar 2;7:43634. doi: 10.1038/srep43634. Sci Rep. 2017. PMID: 28252663 Free PMC article. - Recent zoonoses caused by influenza A viruses.
Alexander DJ, Brown IH. Alexander DJ, et al. Rev Sci Tech. 2000 Apr;19(1):197-225. doi: 10.20506/rst.19.1.1220. Rev Sci Tech. 2000. PMID: 11189716 Review. - Nosocomial influenza in children.
Maltezou HC, Drancourt M. Maltezou HC, et al. J Hosp Infect. 2003 Oct;55(2):83-91. doi: 10.1016/s0195-6701(03)00262-7. J Hosp Infect. 2003. PMID: 14529631 Review.
Cited by
- Unlocking proteomic heterogeneity in complex diseases through visual analytics.
Bhavnani SK, Dang B, Bellala G, Divekar R, Visweswaran S, Brasier AR, Kurosky A. Bhavnani SK, et al. Proteomics. 2015 Apr;15(8):1405-18. doi: 10.1002/pmic.201400451. Proteomics. 2015. PMID: 25684269 Free PMC article. Review. - Objective measures for sentinel surveillance in network epidemiology.
Holme P. Holme P. Phys Rev E. 2018 Aug;98(2-1):022313. doi: 10.1103/PhysRevE.98.022313. Phys Rev E. 2018. PMID: 30253620 Free PMC article. - How Physical Proximity Shapes Complex Social Networks.
Stopczynski A, Pentland A', Lehmann S. Stopczynski A, et al. Sci Rep. 2018 Dec 7;8(1):17722. doi: 10.1038/s41598-018-36116-6. Sci Rep. 2018. PMID: 30531809 Free PMC article. - Optimizing COVID-19 surveillance using historical electronic health records of influenza infections.
Du Z, Bai Y, Wang L, Herrera-Diestra JL, Yuan Z, Guo R, Cowling BJ, Meyers LA, Holme P. Du Z, et al. PNAS Nexus. 2022 Apr 14;1(2):pgac038. doi: 10.1093/pnasnexus/pgac038. eCollection 2022 May. PNAS Nexus. 2022. PMID: 35693630 Free PMC article. - Intimate partner violence norms cluster within households: an observational social network study in rural Honduras.
Shakya HB, Hughes DA, Stafford D, Christakis NA, Fowler JH, Silverman JG. Shakya HB, et al. BMC Public Health. 2016 Mar 8;16:233. doi: 10.1186/s12889-016-2893-4. BMC Public Health. 2016. PMID: 26951919 Free PMC article.
References
- Anonymous. Update: Influenza Activity – United States, August 30, 2009 – January 9, 2010. Morb Mortal Weekly Rep. 2010;59:38–48. - PubMed
- Ginsberg J, Mohebbi MH, Patel RS, Brammer L, Smolinski MS, et al. Detecting influenza epidemics using search engine query data. Nature. 2009;457:1012–1014. - PubMed
- Carneiro HA, Mylonakis E. Google trends: a web-based tool for real-time surveillance of disease outbreaks. Clinical Infect Dis. 2009;49:1557–1564. - PubMed
- Christley RM, Pinchbeck GL, Bowers RG, Clancy D, French NP, et al. Infection in social networks: using network analysis to identify high-risk individuals. Am J Epidemiol. 2005;162:1024–1031. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical