H2A.Z maintenance during mitosis reveals nucleosome shifting on mitotically silenced genes - PubMed (original) (raw)
H2A.Z maintenance during mitosis reveals nucleosome shifting on mitotically silenced genes
Theresa K Kelly et al. Mol Cell. 2010.
Abstract
Profound chromatin changes occur during mitosis to allow for gene silencing and chromosome segregation followed by reactivation of memorized transcription states in daughter cells. Using genome-wide sequencing, we found H2A.Z-containing +1 nucleosomes of active genes shift upstream to occupy TSSs during mitosis, significantly reducing nucleosome-depleted regions. Single-molecule analysis confirmed nucleosome shifting and demonstrated that mitotic shifting is specific to active genes that are silenced during mitosis and, thus, is not seen on promoters, which are silenced by methylation or mitotically expressed genes. Using the GRP78 promoter as a model, we found H3K4 trimethylation is also maintained while other indicators of active chromatin are lost and expression is decreased. These key changes provide a potential mechanism for rapid silencing and reactivation of genes during the cell cycle.
Copyright © 2010 Elsevier Inc. All rights reserved.
Figures
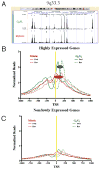
Figure 1. Genome-wide analysis by ChIP-seq shows that H2A.Z localization is maintained during mitosis and preferentially marks +1 nucleosomes of active genes, which shift upstream to occupy the TSS during mitosis, shrinking the NDR
H2A.Z ChIP-seq reads from G0/G1 and M-phase cells were aligned to the Human Genome. (A) Low resolution example demonstrates that global H2A.Z localization is similar during G0/G1 and M-phase. Significant H2A.Z signal is found at the TSSs of expressed (*), but less so at silent (§) genes. In addition, H2A.Z is also present at regions that do not correspond to known TSS (†). (B) Well-positioned +1 nucleosomes of active genes shift upstream to occupy the TSS during mitosis. The size of the shift is promoter specific resulting in a broad peak during M-phase (dashed ovals). (C) Genes, which are not expressed, or expressed at low levels contain little H2A.Z during both M-phase and G0/G1. Forward and reverse reads (solid and dashed lines, respectively) correspond to orientation to the genome. Reads were normalized to the number of promoters analyzed. TSS in b & c are indicated by yellow lines.
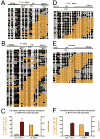
Figure 2. H2A.Z containing nucleosomes upstream and downstream of TSSs of active genes generate a NDR that is shortened during mitosis
(A) Average coverage statistics for each H2A.Z profile covering 1000bp upstream and downstream of known transcription start sites (UCSC). Clusters reveal strong nucleosomal phasing in the G0/G1 data, with nucleosomes detected on one or two sides of a strong NDR at the TSS. The mitotic profiles show a weaker NDR and shifted average position of the +1 and -2 nucleosomes. (B) High resolution examples of nucleosome sliding from cluster 1 (WFS1) and cluster 4 (DDX17) demonstrate promoter specificity in the size of the shift, while the height of the peaks indicate that within an individual promoter the nucleosomes are well-positioned during both G0/G1 and M-phase. p16 (Cluster 7) and MYOD1 (cluster 11), which are silenced by DNA methylation, do not contain H2A.Z at their promoters. (C) Gene expression of representative genes from clusters 1 & 4, WFS1 and DDX17, respectively, show that expression is decreased during mitosis, while genes which do not contain H2A.Z, p16 and MYOD1, are not expressed regardless of cell cycle stage. (D) The size distribution of the NDRs is shown as distance between the -2 and +1 H2A.Z nucleosomes in the G0/G1 (green) and M (red) conditions. The percentage of promoters is plotted along the Y-axis and the distance between the -2 and +1 H2A.Z nucleosomes is on the X-axis. There is clear compaction of the NDR in clusters showing H2A.Z presence during M- phase. (A.U.): Arbitrary units.
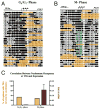
Figure 3. The +1 nucleosomes on the GRP78 and WFS1 promoters shift upstream to occupy the TSS during mitosis, coinciding with decreased gene expression
Black and white circles indicate methylated (accessible) and unmethylated (inaccessible) CpG sites, respectively. Orange highlights M.SssI inaccessible regions that can accommodate a nucleosome (>146 bp). The well-positioned nucleosome after the TSS of GRP78 (A) and WFS1 (D) during G0/G1 shifts upstream to occupy the TSS during mitosis (B, E) in T24 cells. (C, F) Nucleosome occupancy at the TSS is anti-correlated with expression. The percentage of promoter replicas that contained a nucleosome at the TSS (gold bar) is plotted along with GRP78 expression (maroon bar). Expression bars are relative to GAPDH expression and represent the mean + SEM of 3 independent experiments.
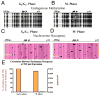
Figure 4. Nucleosome occupancy at TSSs of the mitotically expressed PLK1 promoter does not change during the cell cycle
Black and white circles indicate methylated (accessible) and unmethylated (inaccessible) CpG sites. Orange highlights M.SssI inaccessible regions that can accommodate a nucleosome (>146 bp). Green and blue highlight M.SssI inaccessible regions that mark E2F and TBP binding sites, respectively. The +1 nucleosome of the PLK1 promoter is positioned after the TSS during G0/G1 (A) and M-phase (B). (C) While nucleosome occupancy at the TSS of PLK1 is similar during G0/G1 and M-phase, expression is restricted to M-phase. The percentage of promoter replicas that contained a nucleosome at the TSS (gold bar) is plotted along with PLK1 expression (maroon bar). Expression bars are relative to GAPDH expression and represent the mean + SEM of 3 independent experiments.
Figure 5. Nucleosome occupancy at TSSs of promoters silenced by methylation does not change during the cell cycle
(A, B) p16 is methylated in T24 cells. Black and white circles indicate endogenously methylated and unmethylated CpG sites. (C, D) Black and white circles indicate M.CviPI accessible (methylated) and inaccessible (unmethylated) GpC sites. Pink highlights M.CviPI inaccessible regions that can accommodate a nucleosome (>146 bp). The p16 promoter is inaccessible during G0/G1 (C) and M-phase (D). (E) Nucleosome occupancy is high at the TSS of p16 during G0/G1 and M-phase and expression is not detected. The percentage of promoter replicas that contained a nucleosome at the TSS (gold bar) is plotted along with p16 expression (maroon bar). Expression bars are relative to GAPDH expression and represent the mean + SEM of 3 independent experiments.
Figure 6. The GRP78 promoter has differential nucleosome positioning, unique complements of DNA binding proteins and histone modifications in mitotic and G0/G1 chromatin
(A) H3 ChIP confirms that there is a change in nucleosome occupancy during mitosis. (B) Acetylation of H3 and H2A.Z are decreased during M-phase, relative to G0/G1, while H3K4me3 and H2A.Z are maintained and H3S10 phosphorylation is increased. The extremely low level of H3K27me3 (data not shown) is unchanged during the cell cycle. Bars in (B) indicate ChIP signal from M-phase chromatin (R5.2) expressed as fold change relative to G0/G1 chromatin (R3.3). (C) Pol II, NF-Y and TBP binding is significantly reduced during mitosis. Paired t-test was performed (*= p-value<0.05). Bars represent mean +SEM of at least 6 ChIPs. Arrows in schematic indicate NF-Y and TBP binding sites.
Figure 7. Depiction of Nucleosome Sliding on the GRP78 promoter
During G0/G1, the +1 nucleosome is positioned just after the TSS allowing for TBP, NF-Y and Pol II binding at the TSS. This nucleosome contains the H2A.Z variant, which is acetylated. H3 is acetylated and lysine 4 is tri-methylated. During mitosis the +1 nucleosome shifts to cover the TSS and transcription factor and Pol II binding are lost. The nucleosome loses H3 and H2A.Z acetylation while maintaining the H2A.Z variant and H3K4me3 and acquiring the mitosis specific phosphorylation of serine 10 of H3.
Similar articles
- Histone H2A.Z inheritance during the cell cycle and its impact on promoter organization and dynamics.
Nekrasov M, Amrichova J, Parker BJ, Soboleva TA, Jack C, Williams R, Huttley GA, Tremethick DJ. Nekrasov M, et al. Nat Struct Mol Biol. 2012 Nov;19(11):1076-83. doi: 10.1038/nsmb.2424. Epub 2012 Oct 21. Nat Struct Mol Biol. 2012. PMID: 23085713 - Acetylation of H2A.Z is a key epigenetic modification associated with gene deregulation and epigenetic remodeling in cancer.
Valdés-Mora F, Song JZ, Statham AL, Strbenac D, Robinson MD, Nair SS, Patterson KI, Tremethick DJ, Stirzaker C, Clark SJ. Valdés-Mora F, et al. Genome Res. 2012 Feb;22(2):307-21. doi: 10.1101/gr.118919.110. Epub 2011 Jul 25. Genome Res. 2012. PMID: 21788347 Free PMC article. - Histone variant selectivity at the transcription start site: H2A.Z or H2A.Lap1.
Nekrasov M, Soboleva TA, Jack C, Tremethick DJ. Nekrasov M, et al. Nucleus. 2013 Nov-Dec;4(6):431-8. doi: 10.4161/nucl.26862. Epub 2013 Nov 8. Nucleus. 2013. PMID: 24213378 Free PMC article. - Patterning chromatin: form and function for H2A.Z variant nucleosomes.
Raisner RM, Madhani HD. Raisner RM, et al. Curr Opin Genet Dev. 2006 Apr;16(2):119-24. doi: 10.1016/j.gde.2006.02.005. Epub 2006 Feb 28. Curr Opin Genet Dev. 2006. PMID: 16503125 Review. - The specificity of H2A.Z occupancy in the yeast genome and its relationship to transcription.
Iyer VR. Iyer VR. Curr Genet. 2020 Oct;66(5):939-944. doi: 10.1007/s00294-020-01087-7. Epub 2020 Jun 14. Curr Genet. 2020. PMID: 32537667 Review.
Cited by
- PDGF-AB and 5-Azacytidine induce conversion of somatic cells into tissue-regenerative multipotent stem cells.
Chandrakanthan V, Yeola A, Kwan JC, Oliver RA, Qiao Q, Kang YC, Zarzour P, Beck D, Boelen L, Unnikrishnan A, Villanueva JE, Nunez AC, Knezevic K, Palu C, Nasrallah R, Carnell M, Macmillan A, Whan R, Yu Y, Hardy P, Grey ST, Gladbach A, Delerue F, Ittner L, Mobbs R, Walkley CR, Purton LE, Ward RL, Wong JW, Hesson LB, Walsh W, Pimanda JE. Chandrakanthan V, et al. Proc Natl Acad Sci U S A. 2016 Apr 19;113(16):E2306-15. doi: 10.1073/pnas.1518244113. Epub 2016 Apr 4. Proc Natl Acad Sci U S A. 2016. PMID: 27044077 Free PMC article. - The role of DNA methylation in directing the functional organization of the cancer epigenome.
Lay FD, Liu Y, Kelly TK, Witt H, Farnham PJ, Jones PA, Berman BP. Lay FD, et al. Genome Res. 2015 Apr;25(4):467-77. doi: 10.1101/gr.183368.114. Epub 2015 Mar 6. Genome Res. 2015. PMID: 25747664 Free PMC article. - Binding of NF-κB to nucleosomes: effect of translational positioning, nucleosome remodeling and linker histone H1.
Lone IN, Shukla MS, Charles Richard JL, Peshev ZY, Dimitrov S, Angelov D. Lone IN, et al. PLoS Genet. 2013;9(9):e1003830. doi: 10.1371/journal.pgen.1003830. Epub 2013 Sep 26. PLoS Genet. 2013. PMID: 24086160 Free PMC article. - A 3'UTR polymorphism modulates mRNA stability of the oncogene and drug target Polo-like Kinase 1.
Akdeli N, Riemann K, Westphal J, Hess J, Siffert W, Bachmann HS. Akdeli N, et al. Mol Cancer. 2014 Apr 26;13:87. doi: 10.1186/1476-4598-13-87. Mol Cancer. 2014. PMID: 24767679 Free PMC article. - Crystal structures of heterotypic nucleosomes containing histones H2A.Z and H2A.
Horikoshi N, Arimura Y, Taguchi H, Kurumizaka H. Horikoshi N, et al. Open Biol. 2016 Jun;6(6):160127. doi: 10.1098/rsob.160127. Open Biol. 2016. PMID: 27358293 Free PMC article.
References
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. - PubMed
- Belmont AS. Large-scale chromatin organization. Dordrecht: Kluwer Academic Publishers; 1997.
- Belmont AS. Mitotic chromosome structure and condensation. Curr Opin Cell Biol. 2006;18:632–638. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous