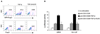Positive and negative transcriptional regulation of the Foxp3 gene is mediated by access and binding of the Smad3 protein to enhancer I - PubMed (original) (raw)
Positive and negative transcriptional regulation of the Foxp3 gene is mediated by access and binding of the Smad3 protein to enhancer I
Lili Xu et al. Immunity. 2010.
Abstract
The molecular mechanisms underlying retinoic acid (RA) augmentation of T cell receptor (TCR) and transforming growth factor-β (TGF-β)-induced Foxp3 transcription and inhibition of the latter by cytokines such as IL-27 were here shown to be related processes involving modifications of baseline (TGF-β-induced) phosphorylated Smad3 (pSmad3) binding to a conserved enhancer region (enhancer I). RA augmentation involved the binding of retinoic acid receptor (RAR) and retinoid X receptor (RXR) to a dominant site in enhancer I and a subordinate site in the promoter. This led to increased histone acetylation in the region of the Smad3 binding site and increased binding of pSmad3. Cytokine (IL-27) inhibition involved binding of pStat3 to a gene silencer in a second conserved enhancer region (enhancer II) downstream from enhancer I; this led to loss of pSmad3 binding to enhancer I. Thus, control of accessibility and binding of pSmad3 provides a common framework for positive and negative regulation of TGF-β-induced Foxp3 transcription.
Copyright © 2010 Elsevier Inc. All rights reserved.
Figures
Figure 1. AP-1 site located in a Foxp3 enhancer region plays positive role on TCR/TGF-β induced Foxp3 gene expression
(A) Murine CD4+ T cells from Foxp3-GFP mice were stimulated with plate bound anti-CD3 (10µg/ml), soluble anti-CD28 (2µg/ml) and TGF-β (5ng/ml) with or without JNK inhibitor (10µM) for 5 days then collected for flow cytometry analysis. (B) Schematic diagram of Foxp3 gene (upper) and AP-1: NFAT and Smad3 sites in Foxp3 enhancer I region (lower). (C) Two different murine T cell lines LBRM (left) and EL4 (right) were transfected with Foxp3 promoter and enhancer I reporter construct (promoter+enhancer I) or the reporter construct with promoter AP-1 site deletion (promoter+enhancer I promoter AP-1 del) or the reporter construct with enhancer I AP-1 site deletion (promoter+enhancer I enhancer AP-1 del). Four hours later cells were split and stimulated with plate bound anti-CD3 (10µg/ml), soluble anti-CD28 (2µg/ml) with or without TGF-β (2ng/ml). Luciferase activity was measured 24 hours later and values were shown as the ratio of firefly luciferase activity/Renilla luciferase activity from the same well. Data represent four independent experiments. **p<0.01, compared to intact construct under same stimulation conditions.
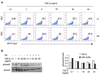
Figure 2. TGF-βRI kinase activity is essential for TCR-/TGF-β-induced Foxp3 expression in murine T cells
(A) Purified CD4+ cells from Foxp3-GFP mice (upper panels) or B6 mice (lower panels) were stimulated with plate-bound anti-CD3 (10µg/ml), soluble anti-CD28 (2µg/ml) and TGF-β (5ng/ml) with or without ALK5 inhibitor for 4 days then subjected for flow cytometry analysis. (B) LBRM or EL4 cells were transfected with Foxp3 promoter and enhancer reporter construct then stimulated with plate bound anti-CD3 (10µg/ml), soluble anti-CD28 (2µg/ml) and TGF-β(2ng/ml) with or without ALK5 inhibitor for 20 hrs. Luciferase activity was measured 24 hours later and values were shown as the ratio of firefly luciferase activity/Renilla luciferase activity from the same well. Data are representative of three independent experiments. **p<0.01, compared to the luciferase activity without ALK5 inhibitor.
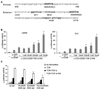
Figure 3. Identification of a Foxp3 silencer containing a Stat3 binding site
(A) Purified CD4+ cells from Stat3 fl/fl and Stat3 fl/fl; MMTV-Cre mice(left panels); Socs3 fl/fl and Socs3 fl/fl; MMTV-Cre mice(right panels) were stimulated with plate bound anti-CD3 (10µg/ml), soluble anti-CD28 (2µg/ml) and TGF-β(5ng/ml) with or without IL-27 (20ng/ml) for 4 days then collected for Foxp3 staining and subjected to flow cytometry analysis. (B) Alignment of human and mouse Foxp3 enhancer II region containing stat3 binding site(up) and the structure of Foxp3 promoter and enhancer I and enhancer II (contains stat3 binding site) reporter construct. (C) Murine T cell line LBRM were transfected with Foxp3 promoter and enhancer I reporter construct or Foxp3 promoter and Enhancer I and enhancer II reporter construct or Foxp3 promoter and Enhancer I and enhancer II reporter construct with stat3 binding site deletion then stimulated with plate bound anti-CD3 (10µg/ml), soluble anti-CD28 (2µg/ml) and TGF-β (2ng/ml). Luciferase activity was measured 24 hours later and values were shown as the ratio of firefly luciferase activity/Renilla luciferase activity from the same well. Data are representative of four independent experiments. *p<0.05, **p<0.01, compared to intact construct under same stimulation conditions.
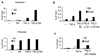
Figure 4. The positive effect of retinoic acid on Foxp3 expression completely depends on intact TGF-βRI kinase activity and Smad3
(A) Purified CD4+ cells from Foxp3-GFP mice were stimulated with plate bound anti-CD3 (10µg/ml), soluble anti-CD28 (2µg/ml) and TGF-β (5ng/ml) with or without retinoic acid (100nM) and JNK kinase inhibitor (10µM), cyclosporine A (20nM), IL-27 (20ng/ml) or ALK5 inhibitor (5µM) for 4 days then collected for flow cytometry analysis. (B) Purified CD4+ cells from B6-WT littermate mice and B6-Smad3 knockout were stimulated with plate-bound anti-CD3 (10µg/ml), soluble anti-CD28 (1µg/ml), TGF-β(5ng/ml), RA (100nM) with or without ALK5 inhibitor (2.5µM) for 4 days. Cells were collected and stained with APC anti-Foxp3 (eBioscience) then subjected to flow cytometric analysis.
Figure 5. The enhancement of RA on TCR/TGF-β induced Foxp3 transcription is not due to increased Smad3 phosphorylation
(A) Purified CD4+ cells from Foxp3-GFP mice was stimulated with plate bound anti-CD3 (10µg/ml), soluble anti-CD28 (2µg/ml) and TGF-β at different concentrations as indicated with or without retinoic acid (200nM) for 4 days then collected for flow cytometry analysis. (B) Purified CD4+ cells from B6 mice were stimulated with TGF-β at different concentrations as indicated with or without retinoic acid (500nM) for 2 hours. Cells were collected and cell lysates were prepared and subjected to Western-Blot (left). Intensity of bands was measured using NIH Image software and showed as the ratio between p-Smad3 and Smad3 from the same sample (right). Data are representative of two independent experiments.
Figure 6. Retinoic acid directly regulates Foxp3 promoter and enhancer activity
(A) Location of two RAR-RXR binding sites in Foxp3 promoter and enhancer I regions. (B) LBRM (left) or EL4 (right) cells were transfected with a Foxp3 promoter and enhancer I reporter construct and stimulated with plate bound anti-CD3 (10µg/ml), soluble anti-CD28 (2µg/ml) plus TGF-β (2ng/ml) and retinoic acid at a series of concentration as indicated. Luciferase activity was measured 24 hours later and values were shown as the ratio of firefly luciferase activity/renilla luciferase activity from the same well. Data are representative of two independent experiments. **p<0.01, compared to the luciferase activity without RA. (C) EL4 cells were transfected with Foxp3 promoter and enhancer I reporter construct with deletion of the RAR-RXR binding site in either the promoter region or enhancer region (as indicated in A) or both and then stimulated with plate-bound anti-CD3 (10µg/ml), soluble anti-CD28 (2µg/ml) and TGF-β (2ng/ml) with or without retinoic acid (500nM) as indicated. (ProRAR-RXR del: promoter RAR-RXR deletion; EnhRAR-RXR del: enhancer I RAR-RXR deletion; Pro+Enh RAR-RXR del: promoter+enhancer I RAR-RXR deletion). Luciferase activity was measured 24 hours later and values were shown as the ratio of firefly luciferase activity/Renilla luciferase activity from the same well. Data are representative of three independent experiments. **p<0.01, compared to intact construct under same stimulation conditions.
Figure 7. RA increases histone acetylation at the Enhancer region containing NF-AT/Smad3 binding sites and facilitates increased Smad3 binding to Enhancer
(A) Purified CD4+ cells from B6 mice were stimulated with plate bound anti-CD3 (10µg/ml), soluble anti-CD28 (2µg/ml) and TGF-β (5ng/ml) with or without retinoic acid (200nM) for 48 hours. CHIP assay was performed using anti-acetyi histone H4 antibody or rabbit IgG. Precipitated chromatins were subjected to real time PCR using primers targeting enhancerI region (upper) or promoter region (lower). Values were shown as the percentage of corresponding input. Data are representative of three independent experiments. (B) Purified CD4+ cells from B6 mice were treated as in (A) with or without IL-27 (20ng/ml) for 2 hours. CHIP assay was performed using anti-Smad3 antibody or rabbit IgG. Precipitated chromatins were subjected to real time PCR using primers targeting enhancer I region. Values were shown as the percentage of corresponding input. Data are representative of two independent experiments. (C) Purified CD4+ cells from B6 mice were stimulated with plate bound anti-CD3 (10µg/ml), soluble anti-CD28 (2µg/ml) and TGF-β (5ng/ml) with or without JNK kinase inhibitor (10µM) for 2 hours. CHIP assay was performed using anti-Smad3 antibody or rabbit IgG as described in (B).
Similar articles
- Smad3 and NFAT cooperate to induce Foxp3 expression through its enhancer.
Tone Y, Furuuchi K, Kojima Y, Tykocinski ML, Greene MI, Tone M. Tone Y, et al. Nat Immunol. 2008 Feb;9(2):194-202. doi: 10.1038/ni1549. Epub 2007 Dec 23. Nat Immunol. 2008. PMID: 18157133 - Retinoic acid-induced CCR9 expression requires transient TCR stimulation and cooperativity between NFATc2 and the retinoic acid receptor/retinoid X receptor complex.
Ohoka Y, Yokota A, Takeuchi H, Maeda N, Iwata M. Ohoka Y, et al. J Immunol. 2011 Jan 15;186(2):733-44. doi: 10.4049/jimmunol.1000913. Epub 2010 Dec 8. J Immunol. 2011. PMID: 21148038 - Transforming growth factor-beta inhibits pulmonary surfactant protein B gene transcription through SMAD3 interactions with NKX2.1 and HNF-3 transcription factors.
Li C, Zhu NL, Tan RC, Ballard PL, Derynck R, Minoo P. Li C, et al. J Biol Chem. 2002 Oct 11;277(41):38399-408. doi: 10.1074/jbc.M203188200. Epub 2002 Aug 2. J Biol Chem. 2002. PMID: 12161428 - What turns on Foxp3?
von Boehmer H, Nolting J. von Boehmer H, et al. Nat Immunol. 2008 Feb;9(2):121-2. doi: 10.1038/ni0208-121. Nat Immunol. 2008. PMID: 18204423 No abstract available. - TGF-β-mediated Foxp3 gene expression is cooperatively regulated by Stat5, Creb, and AP-1 through CNS2.
Ogawa C, Tone Y, Tsuda M, Peter C, Waldmann H, Tone M. Ogawa C, et al. J Immunol. 2014 Jan 1;192(1):475-83. doi: 10.4049/jimmunol.1301892. Epub 2013 Dec 2. J Immunol. 2014. PMID: 24298014 Free PMC article.
Cited by
- Regulatory T cells: mechanisms of differentiation and function.
Josefowicz SZ, Lu LF, Rudensky AY. Josefowicz SZ, et al. Annu Rev Immunol. 2012;30:531-64. doi: 10.1146/annurev.immunol.25.022106.141623. Epub 2012 Jan 6. Annu Rev Immunol. 2012. PMID: 22224781 Free PMC article. Review. - FOXP3 expression is modulated by TGF‑β1/NOTCH1 pathway in human melanoma.
Skarmoutsou E, Bevelacqua V, D' Amico F, Russo A, Spandidos DA, Scalisi A, Malaponte G, Guarneri C. Skarmoutsou E, et al. Int J Mol Med. 2018 Jul;42(1):392-404. doi: 10.3892/ijmm.2018.3618. Epub 2018 Apr 4. Int J Mol Med. 2018. PMID: 29620159 Free PMC article. - Helper T cell plasticity: impact of extrinsic and intrinsic signals on transcriptomes and epigenomes.
Bonelli M, Shih HY, Hirahara K, Singelton K, Laurence A, Poholek A, Hand T, Mikami Y, Vahedi G, Kanno Y, O'Shea JJ. Bonelli M, et al. Curr Top Microbiol Immunol. 2014;381:279-326. doi: 10.1007/82_2014_371. Curr Top Microbiol Immunol. 2014. PMID: 24831346 Free PMC article. Review. - Tumor growth limited to subcutaneous site vs tumor growth in pulmonary site exhibit differential effects on systemic immunities.
Masuda J, Takayama E, Strober W, Satoh A, Morimoto Y, Honjo Y, Ichinohe T, Tokuno SI, Ishizuka T, Nakata T, Mizutani A, Umemura N, Kitani A, Fuss IJ, Shigehiro T, Kawaki H, Mizuno-Kamiya M, Kondoh N, Seno M. Masuda J, et al. Oncol Rep. 2017 Jul;38(1):449-455. doi: 10.3892/or.2017.5646. Epub 2017 May 17. Oncol Rep. 2017. PMID: 28535011 Free PMC article. - SMAD regulatory networks construct a balanced immune system.
Malhotra N, Kang J. Malhotra N, et al. Immunology. 2013 May;139(1):1-10. doi: 10.1111/imm.12076. Immunology. 2013. PMID: 23347175 Free PMC article. Review.
References
- Derynck R, Zhang YE. Smad-dependent and Smad-independent pathways in TGF-beta family signalling. Nature. 2003;425:577–584. - PubMed
- Huber M, Steinwald V, Guralnik A, Brüstle A, Kleemann P, Rosenplänter C, Decker T, Lohoff M. IL-27 inhibits the development of regulatory T cells via STAT3. Int Immunol. 2008;20:223–234. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases