Methylation of the retinoblastoma tumor suppressor by SMYD2 - PubMed (original) (raw)
Methylation of the retinoblastoma tumor suppressor by SMYD2
Louis A Saddic et al. J Biol Chem. 2010.
Abstract
The retinoblastoma tumor suppressor (RB) is a central cell cycle regulator and tumor suppressor. RB cellular functions are known to be regulated by a diversity of post-translational modifications such as phosphorylation and acetylation, raising the possibility that RB may also be methylated in cells. Here we demonstrate that RB can be methylated by SMYD2 at lysine 860, a highly conserved and novel site of modification. This methylation event occurs in vitro and in cells, and it is regulated during cell cycle progression, cellular differentiation, and in response to DNA damage. Furthermore, we show that RB monomethylation at lysine 860 provides a direct binding site for the methyl-binding domain of the transcriptional repressor L3MBTL1. These results support the idea that a code of post-translational modifications exists for RB and helps guide its functions in mammalian cells.
Figures
FIGURE 1.
Methylation of human RB by SMYD2 at lysine 860 in vitro. A, schematic representation showing RB N-terminal region (N), its pocket (A, I, B), and its C-terminal domain. Known acetylated (Ac) lysines (positions 873 and 874) and the lysine methylated by SMYD2 (position 860) are shown. B, RB is methylated by SMYD2 at lysine 860 in vitro. Autoradiograms (3H) of histone methyltransferase (HMTase) assays with recombinant RB fragments (GST-RB) and SMYD2. Single amino acid substitutions (K-R) are indicated. Coomassie Blue staining shows the amount of RB protein loaded. C, C-term RB methylation is specific to SMYD2. Autoradiogram of an HMTase assay with GST-RB fragments and SET9. D, RB lysine 860 is highly conserved. Conservation of the sequences surrounding RB lysine 860 (in red) in several species.
FIGURE 2.
Mass spectrometry analysis of RB methylation by SMYD2 in vitro. A, RB is monomethylated by SMYD2. Modified peptides identified, charge state, and highest scoring site of modification (degree of methylation) in the RB/SMYD2 mass spectrometry analysis are shown. Lysine 860 is bolded and underlined. B, electron transfer dissociation (ETD) MS/MS spectrum matching charge state 6+ for the peptide ILVSIGESFGTSEKFQKINQMVCNSDRVLK(Me)R and corresponding fragmentation map. Matched c- and z-ions are indicated on the fragmentation map along with their m/z values. Matched c- and z-ions as well as charge-reduced precursors are labeled on the MS/MS spectrum. Lysine monomethylation can be localized to Lys-860 and not Lys-844 or Lys-847 based on site-determining ions.
FIGURE 3.
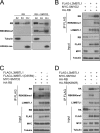
Methylation of human RB by SMYD2 at lysine 860 in vivo. A, RBK860me1 antibody recognizes monomethyl Lys-860 RB. The specificity of polyclonal antibodies recognizing monomethylated Lys-860 in the RB protein (RBK860me1) was assessed by dot blot analysis with biotinylated peptides. A peptide from histone H4 (amino acids 1–20) serves as a negative control. Streptavidin was used as a positive control and a loading control. B, overexpressed SMYD2 monomethylates overexpressed RB at lysine 860 in 293T cells. Whole cell extracts from 293T cells transfected with the indicated plasmids were immunoprecipitated with HA resin and analyzed by immunoblot with RB, RBK860me1, and E2F antibodies. Controls include lysine to arginine substitutions in RB at Lys-860 and Lys-870, -873, -874 as well as a mutant form of SMYD2 (Y290F) with decreased methyltransferase activity. Inputs for HA-RB and MYC-SMYD2 are shown. C, overexpressed SMYD2 monomethylates endogenous RB at lysine 860. Whole cell extracts of 293T cells were immunoprecipitated with antibodies against RB or control IgG antibodies followed by immunoblot analysis with RB and RBK860me1 antibodies. Inputs for RB and MYC-SMYD2 are shown. D, methylation of endogenous RB at K860 in U2OS cells is decreased upon knockdown of SMYD2. shRNA molecules against SMYD2 (shSMYD2) were stably expressed from a lentiviral vector. Cells infected with an empty lentivirus serve as a control. Whole cell extracts were immunoprecipitated with antibodies against RBK860me1 or control IgG antibodies followed by immunoblot analysis with RB. Inputs for RB and actin are shown. The efficiency of the knockdown was measured by quantitative RT-PCR. One representative quantification of the knockdown is indicated.
FIGURE 4.
Regulation of RB methylation at lysine 860 in vivo. A, methylation of RB accumulates in G0. Quiescent T98G cells were stimulated with 20% serum and RB methylation was measured by immunoprecipitation of whole cell extracts with RBK860me1 antibody and immunoblot analysis with RB antibody. Note that RB becomes hyperphosphorylated as cells re-enter the cell cycle, and that the levels of the RB/E2F target p107 increase as cells progress into S phase. Actin serves as a loading control. Propidium iodide FACS profiles (bottom) show cell cycle re-entry. B, methylated RB accumulates as C2C12 myoblasts differentiate. Whole cell extracts from differentiating C2C12 myoblasts were immunoprecipitated with RBK860me1 antibody followed by immunoblot for RB. Input levels of RB are shown along with actin as a loading control (top). A replicate of this experiment is shown with RB loading equalized from day 0 to day 2 (middle). Photographs at days 0 and 2 show the differentiation into myotubes (bottom). C, RB methylation accumulates with DNA damage. Whole cell extracts from NIH3T3 cells treated with etoposide for the indicated time periods were immunoprecipitated with RBK860me1 followed by immunoblot analysis. Immunoblotting input with the phospho-H2AX epitope (γH2AX) serves as a positive control for the accumulation of DNA damage. Input levels of RB are shown along with actin as a loading control.
FIGURE 5.
RB methylation at Lys-860 increases its interaction with the MBT domains of L3MBTL1 in vitro. A, 3xMBT from L3MBTL1 preferentially binds mono- and dimethyl K860 RB fragments. Pull-down assays of biotinylated RB and H4 peptides containing varying degrees of methylation with GST-tagged 3xMBT from L3MBTL1. B, mutations in key residues of the methyl-binding pocket of 3xMBT L3MBTL1 reduce binding to methylated RB fragments. Pull-down assays of biotinylated RBK860me0 and RBK860me1 peptides with 3xMBT L3MBTL1 mutants. Input levels of the RB fragment are shown to the left of each blot. C and D, calorimetry assays confirm and quantify binding of methylated RB to 3xMBT L3MBTL1. Isothermal titration calorimetry assays were performed to measure the affinity of the interaction between RB and the 3xMBT domain of L3MBTL1. Only the interaction with the monomethylated RB peptide is shown in C. All the data are quantified in D; no Kd could be determined for K860me0 and K860me3 because of the low affinity of the interaction.
FIGURE 6.
RB methylation at Lys-860 increases its interaction with the MBT domains of L3MBTL1 in vivo. A, monomethyl Lys-860 RB can be localized to chromatin. 293T cells were fractionated by detergent lysis to generate a cytoplasmic S2 fraction, a nuclear S3 fraction, and a chromatin (Chr) fraction. Fractions were then analyzed by immunoblot with antibodies against RB and RBK860me1. Tubulin and H3K4tri-me were used to assess the purity of the fractionation. B, overexpression of SMYD2 increases binding of RB to L3MBTL1. 293T cells were transfected with the indicated expression plasmids. Whole cell extracts were immunoprecipitated with FLAG resin and analyzed by immunoblot with antibodies against RB, RBK860me1, and L3MBTL1. Inputs for FLAG-L3MBTL1, MYC-SMYD2, and RB are shown. C, D355N mutation in the methyl-binding pocket of L3MBTL1 reduces the affinity of L3MBTL1 for RB. 293T cells were transfected with the indicated expression plasmids. IP was performed as in B and analyzed by immunoblot with antibodies against RB, RBK860me1, and L3MBTL1. Inputs for FLAG-L3MBTL1, FLAG-L3MBTL1(D355N), RB, and MYC-SMYD2 are shown. D, RB(K860R) mutation reduces the affinity of RB for L3MBTL1. 293T cells were transfected with the indicated expression plasmids. Immunoprecipitation was performed as in B and C and analyzed by immunoblot with antibodies against HA, RBK860me1, and L3MBTL1. Inputs for FLAG-L3MBTL1, HA-RB, HA-860, and MYC-SMYD2 are shown. Tubulin serves as a loading control.
Similar articles
- RB1 methylation by SMYD2 enhances cell cycle progression through an increase of RB1 phosphorylation.
Cho HS, Hayami S, Toyokawa G, Maejima K, Yamane Y, Suzuki T, Dohmae N, Kogure M, Kang D, Neal DE, Ponder BA, Yamaue H, Nakamura Y, Hamamoto R. Cho HS, et al. Neoplasia. 2012 Jun;14(6):476-86. doi: 10.1593/neo.12656. Neoplasia. 2012. PMID: 22787429 Free PMC article. - Lysine methyltransferase Smyd2 regulates Hsp90-mediated protection of the sarcomeric titin springs and cardiac function.
Voelkel T, Andresen C, Unger A, Just S, Rottbauer W, Linke WA. Voelkel T, et al. Biochim Biophys Acta. 2013 Apr;1833(4):812-22. doi: 10.1016/j.bbamcr.2012.09.012. Epub 2012 Oct 6. Biochim Biophys Acta. 2013. PMID: 23047121 - The MBT repeats of L3MBTL1 link SET8-mediated p53 methylation at lysine 382 to target gene repression.
West LE, Roy S, Lachmi-Weiner K, Hayashi R, Shi X, Appella E, Kutateladze TG, Gozani O. West LE, et al. J Biol Chem. 2010 Nov 26;285(48):37725-32. doi: 10.1074/jbc.M110.139527. Epub 2010 Sep 24. J Biol Chem. 2010. PMID: 20870725 Free PMC article. - Unsafe SETs: histone lysine methyltransferases and cancer.
Schneider R, Bannister AJ, Kouzarides T. Schneider R, et al. Trends Biochem Sci. 2002 Aug;27(8):396-402. doi: 10.1016/s0968-0004(02)02141-2. Trends Biochem Sci. 2002. PMID: 12151224 Review. - Histone methylation in transcriptional control.
Kouzarides T. Kouzarides T. Curr Opin Genet Dev. 2002 Apr;12(2):198-209. doi: 10.1016/s0959-437x(02)00287-3. Curr Opin Genet Dev. 2002. PMID: 11893494 Review.
Cited by
- Genetic alterations of histone lysine methyltransferases and their significance in breast cancer.
Liu L, Kimball S, Liu H, Holowatyj A, Yang ZQ. Liu L, et al. Oncotarget. 2015 Feb 10;6(4):2466-82. doi: 10.18632/oncotarget.2967. Oncotarget. 2015. PMID: 25537518 Free PMC article. - Structure and function of SET and MYND domain-containing proteins.
Spellmon N, Holcomb J, Trescott L, Sirinupong N, Yang Z. Spellmon N, et al. Int J Mol Sci. 2015 Jan 8;16(1):1406-28. doi: 10.3390/ijms16011406. Int J Mol Sci. 2015. PMID: 25580534 Free PMC article. Review. - Transcriptional regulation by methyltransferases and their role in the heart: highlighting novel emerging functionality.
Szulik MW, Davis K, Bakhtina A, Azarcon P, Bia R, Horiuchi E, Franklin S. Szulik MW, et al. Am J Physiol Heart Circ Physiol. 2020 Oct 1;319(4):H847-H865. doi: 10.1152/ajpheart.00382.2020. Epub 2020 Aug 21. Am J Physiol Heart Circ Physiol. 2020. PMID: 32822544 Free PMC article. Review. - G1 arrest and differentiation can occur independently of Rb family function.
Wirt SE, Adler AS, Gebala V, Weimann JM, Schaffer BE, Saddic LA, Viatour P, Vogel H, Chang HY, Meissner A, Sage J. Wirt SE, et al. J Cell Biol. 2010 Nov 15;191(4):809-25. doi: 10.1083/jcb.201003048. Epub 2010 Nov 8. J Cell Biol. 2010. PMID: 21059851 Free PMC article. - An unexpected journey: lysine methylation across the proteome.
Moore KE, Gozani O. Moore KE, et al. Biochim Biophys Acta. 2014 Dec;1839(12):1395-403. doi: 10.1016/j.bbagrm.2014.02.008. Epub 2014 Feb 20. Biochim Biophys Acta. 2014. PMID: 24561874 Free PMC article. Review.
References
- Sherr C. J., McCormick F. (2002) Cancer Cell 2, 103–112 - PubMed
- Weinberg R. A. (1995) Cell 81, 323–330 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- T32 CA009302/CA/NCI NIH HHS/United States
- R01 CA132685/CA/NCI NIH HHS/United States
- P41 RR011823/RR/NCRR NIH HHS/United States
- F30 AG034741/AG/NIA NIH HHS/United States
- T32 CA009523/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials