Cooperation of breast cancer proteins PALB2 and piccolo BRCA2 in stimulating homologous recombination - PubMed (original) (raw)
. 2010 Oct;17(10):1247-54.
doi: 10.1038/nsmb.1915. Epub 2010 Sep 26.
Affiliations
- PMID: 20871615
- PMCID: PMC4094107
- DOI: 10.1038/nsmb.1915
Cooperation of breast cancer proteins PALB2 and piccolo BRCA2 in stimulating homologous recombination
Rémi Buisson et al. Nat Struct Mol Biol. 2010 Oct.
Abstract
Inherited mutations in human PALB2 are associated with a predisposition to breast and pancreatic cancers. PALB2's tumor-suppressing effect is thought to be based on its ability to facilitate BRCA2's function in homologous recombination. However, the biochemical properties of PALB2 are unknown. Here we show that human PALB2 binds DNA, preferentially D-loop structures, and directly interacts with the RAD51 recombinase to stimulate strand invasion, a vital step of homologous recombination. This stimulation occurs through reinforcing biochemical mechanisms, as PALB2 alleviates inhibition by RPA and stabilizes the RAD51 filament. Moreover, PALB2 can function synergistically with a BRCA2 chimera (termed piccolo, or piBRCA2) to further promote strand invasion. Finally, we show that PALB2-deficient cells are sensitive to PARP inhibitors. Our studies provide the first biochemical insights into PALB2's function with piBRCA2 as a mediator of homologous recombination in DNA double-strand break repair.
Figures
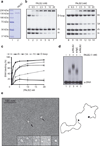
Figure 1. Purified PALB2 binds DNA
(a) SDS-PAGE of purified PALB2 protein. Left: lane 1, Precision plus molecular weight markers (BioRad); lane 2, purified PALB2 (500 ng). (b) PALB2 binds D-loops preferentially. Competitions electrophoretic mobility shift assays were performed with PALB2 (0.5–20 nM) and D-loop, dsDNA (ds), and ssDNA (ss) (lanes 2–7) or Holliday junctions (HJ), splayed arms (SA), dsDNA and ssDNA substrates (lanes 9–14) on 8% acrylamide gel. Lanes 1 and 8, controls without protein. (c) Quantification of electrophoretic mobility shift assays. (d) PALB2-ssDNA protein complexes are supershifted with a PALB2 antibody (lane 3) or a control IgG (lane 4) and are analyzed on 0.8% agarose gel. Lane 1 and 5, controls with DNA alone or incubated with the PALB2 antibody respectively. (e) Electron microscopy of complexes made by PALB2 (40 nM) on a linear DNA molecule (~3900 bp) containing 3’single-strand tails at each end (~200 nt). Insert : closeup view of PALB2 in the absence of DNA. 86% of the DNA molecules observed had both ends bound while 14% had one end bound.
Figure 2. PALB2 stimulates RAD51 strand exchange and DNA capture
(a) PALB2 stimulates RAD51-mediated strand exchange. RAD51 and /or PALB2 were mixed with 1.5 µM 63-mer ssDNA. DNA strand exchange was initiated by addition of homologous 3 µM of 63-bp dsDNA in which the strand that would be displaced during DNA strand exchange was 32P-labeled. (b) Left: scheme of the second capture strategy; right: second capture assays with PALB2 and /or RAD51. Indicated concentration of PALB2 or/and RAD51 were incubated with beads containing 1 µM of 5’-biotinylated ssDNA poly dT. Then 300 nM of non homologous dsDNA was added with the DNA-protein complex. Beads and supernatant were separated, washed and treated with SDS to elute the bound proteins and analyzed by polyacrylamide gel.
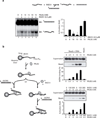
Figure 3. PALB2 interacts directly with RAD51
(a) Right: Scheme of the PALB2 deletion variants fused to GST and the RAD51 GST fusions. Left: and SDS-PAGE of the corresponding purified proteins (b) GST alone or GST-tagged PALB2 fragments purified from bacteria were incubated with RAD51 and glutathione sepharose beads were used to capture protein complexes. The beads were washed and treated with SDS to elute the bound proteins, and revealed by western blotting against RAD51. (c) GST alone or GST-tagged RAD51 fragments were incubated with/without PALB2 followed by immunoprecipitation analysis using PALB2 antibodies and revealed by western blotting against PALB2 and GST. (d) Co-immunoprecipitations of purified PALB2 truncations (P2T1 or P2T5) and RAD51 truncation (R51T3) were performed with anti-IgG or anti-Histidine tag antibodies followed by western blotting using an anti-GST antibody. (e) Co-immunoprecipitation of purified PALB2 and RAD51. Immunoprecipitations were conducted with a polyclonal antibody against PALB2 and blotted against PALB2 and RAD51 as indicated.
Figure 4. PALB2 binds DNA by two distinct domains and stimulates RAD51-mediated D-loop formation
(a) Competitions electrophoretic mobility shift assays were performed with GST-cleaved PALB2 truncations (300 nM) and D-loop, dsDNA (ds), and ssDNA (ss) (lanes 2–6) or Holliday junctions (HJ), splayed arms (SA), dsDNA and ssDNA substrates (lanes 9–13) on 8% acrylamide gel. (b) Electron microscopy of complexes made by PALB2 truncation P2T1 and P2T3 (40 nM) on linear DNA molecule containing 3’single-strand tails of ~200 nucleotides at each end. (c,d) D-loop reactions mediated by RAD51 (300 nM (c) or 400 nM (d)) or PALB2 alone (lanes 2–3), or combinations of RAD51 and PALB2 (lanes 4 to 8) with (a) 1 µM of 100-mer single-stand DNA oligonucleotide. Bottom: quantification of the results. (d) D-loop reactions with 1 µM of linear DNA molecule containing 3’-single-strand tails of ~200 nucleotides at each end. Bottom: quantification of the results. The band indicated with an asterisk corresponds to annealed tailed molecules.
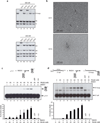
Figure 5. A BRCA2 chimera protein (piccolo BRCA2) recapitulates human BRCA2 properties
(a) The BRCA2 chimera contains the PALB2 binding domain (amino acids 1-40), BRC3 and BRC4 repeats (amino acids 1409-1596), the three OB-folds (amino acids 2477 to 3194), and TR2 repeat (amino acids 3265 to 3330). (b) piBRCA2 colocalizes with γ-H2AX and PALB2 to a unique I-SceI DNA double-strand break. DR95 cells containing DR-GFP were transfected with pCBASce (encoding I-SceI) and immunofluorescence was conducted with the indicated antibodies. All piBRCA2 transfected cells showed co-localization with γ-H2AX. (c-d) piBRCA2 interacts with PALB2 and RAD51. (c) HEK293T cells were transfected with a plasmid expressing piBRCA2-Flag or (d) plasmids expressing piBRCA2-Flag and PALB2-Flag or PALB2 lacking the interaction domain with BRCA2 (WD40 repeat). Whole cell extracts were prepared and immunoprecipitations were conducted using IgG alone, anti-flag or anti-PALB2 antibody, followed by western blotting with the indicated antibodies. (e) The nuclear localization of RAD51 is restored in presence of piBRCA2. Hela cells were transfected with a siRNA againt BRCA2 and with or without a plasmid expressing piBRCA2-Flag. Cells were fractionated into cytosolic and chromatin fractions and analyzed by Western blotting for the presence of piBRCA2 and endogenous BRCA2 and RAD51. Immunoblots against GAPDH and Histone H3 were positive controls for cytosolic and chromatin fractions respectively. (f) piBRCA2 overcomes RPA inhibition to promote RAD51 assembly. RPA bound to a ssDNA oligonucleotide prevents RAD51 assembly (lane 2) whereas addition of piBRCA2 stimulates RAD51 filament formation in presence of RPA (lanes 3 to 7). The stimulation was weaker in absence of ATP and Mg2+ (lane 8). Lanes 9–11, interaction of RPA, RAD51 and PALB2 on beads alone.
Figure 6. A BRCA2 chimera stimulates RAD51-mediated D-loop formation and function with PALB2 in a synergistic manner to promote D-loop formation
(a) SDS-PAGE of the purified piBRCA2 protein (300 ng). Left: lane 1, Precision plus molecular weight markers (BioRad); lane 2, purified piBRCA2. (b) Electrophoretic mobility shift assays were performed with piBRCA2 (0.5–20 nM) and D-loop, dsDNA (ds), and ssDNA (ss) (lanes 2–7) or Holliday junctions (HJ), splayed arms (SA), dsDNA (ds) and ssDNA (ss) substrates (lanes 9–14) and migrated on a 8% acrylamide gel. Lanes 1 and 8, controls without protein. Far right : quantification of percentage of DNA binding. (c) Effect of piBRCA2 in RAD51-catalyzed D-loop formation. Reactions contained DNA alone (lane 1); piBRCA2 (70 nM, lane 2); RAD51 (400 nM, lane 3); or a combination of both proteins (lanes 4–10) with 1 µM on linear DNA molecule containing 3’single-strand tails. Right: Quantification of the results (d) PALB2 (30–40 nM) works in concert with piBRCA2 (15–20 nM) to stimulate RAD51 (400 nM) in D-loop reactions. Right: Quantification of the results. The band indicated with an asterisk in panels c and d corresponds to annealed tailed molecules.
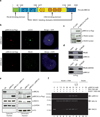
Figure 7. PALB2 is a HR mediator protecting the RAD51 filament from disassembly
(a) RPA bound to a ssDNA oligonucleotide prevents RAD51 assembly (lane 2) whereas addition of PALB2 stimulates RAD51 filament formation in presence of RPA (lanes 3 to 7) but not in absence of ATP and Mg2+ (lane 8). Lanes 9–11, interaction of RPA, RAD51 and PALB2 on beads alone (b) Left, SDS-PAGE of purified BRC3/4-GST polypeptide (1 µg) corresponding to the BRC3 and BRC4 region of BRCA2 (amino acids 1409-1596). Lane 1, Precision plus molecular weight markers (BioRad); lane 2, purified BRC3/4-GST (1 µg). Right, inhibition of RAD51–DNA complex formation by BRC3/4-GST polypeptide. Purified RAD51 protein was incubated with a 100-mer single-strand DNA oligonucleotide (lane 4) prior to the addition of BRC3/4-GST (lanes 5–8). Lane 3, DNA alone; lane 9, BRC3/4-GST incubated with DNA. (c) PALB2 or piBRCA2 protect D-loop formation from a BRC3/4-GST polypeptide. RPA (lanes 5 to 8), PALB2 (lanes 9 to 12) and piBRCA2 (lanes 13 to 16) were incubated with RAD51-3’tail DNA complex prior addition of BRC3/4-GST polypeptide. The band indicated with an asterisk corresponds to annealed tailed molecules. (d) Relative D-loop formation in presence of RPA, PALB2 or piBRCA2 with BRC3/4-GST polypeptide. (e) Survival curves of wild-type lymphoblasts (FEN5280), EUFA1341 (expressing PALB2 Y551X), and EUFA1341 complemented with vector alone (pOZC) or PALB2 to PARP inhibitor (AZD2281). Cell survival was analyzed by quantification of ATP levels.
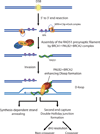
Figure 8
Model. PALB2, and BRCA2 work synergistically to stimulate homologous recombination. Following DNA damage and resection by MRN/CtIP, PALB2-BRCA2 activates RAD51 to promote the invasion of an undamaged template, leading to synthesis-dependant strand annealing or second end capture and double Holliday junction formation to allow DSB repair.
Similar articles
- Enhancement of RAD51 recombinase activity by the tumor suppressor PALB2.
Dray E, Etchin J, Wiese C, Saro D, Williams GJ, Hammel M, Yu X, Galkin VE, Liu D, Tsai MS, Sy SM, Schild D, Egelman E, Chen J, Sung P. Dray E, et al. Nat Struct Mol Biol. 2010 Oct;17(10):1255-9. doi: 10.1038/nsmb.1916. Epub 2010 Sep 26. Nat Struct Mol Biol. 2010. PMID: 20871616 Free PMC article. - Breast cancer-associated missense mutants of the PALB2 WD40 domain, which directly binds RAD51C, RAD51 and BRCA2, disrupt DNA repair.
Park JY, Singh TR, Nassar N, Zhang F, Freund M, Hanenberg H, Meetei AR, Andreassen PR. Park JY, et al. Oncogene. 2014 Oct 2;33(40):4803-12. doi: 10.1038/onc.2013.421. Epub 2013 Oct 21. Oncogene. 2014. PMID: 24141787 Free PMC article. - Human BRCA2 protein promotes RAD51 filament formation on RPA-covered single-stranded DNA.
Liu J, Doty T, Gibson B, Heyer WD. Liu J, et al. Nat Struct Mol Biol. 2010 Oct;17(10):1260-2. doi: 10.1038/nsmb.1904. Epub 2010 Aug 22. Nat Struct Mol Biol. 2010. PMID: 20729859 Free PMC article. - PALB2: the hub of a network of tumor suppressors involved in DNA damage responses.
Park JY, Zhang F, Andreassen PR. Park JY, et al. Biochim Biophys Acta. 2014 Aug;1846(1):263-75. doi: 10.1016/j.bbcan.2014.06.003. Epub 2014 Jul 3. Biochim Biophys Acta. 2014. PMID: 24998779 Free PMC article. Review. - BRCA1-Dependent and Independent Recruitment of PALB2-BRCA2-RAD51 in the DNA Damage Response and Cancer.
Foo TK, Xia B. Foo TK, et al. Cancer Res. 2022 Sep 16;82(18):3191-3197. doi: 10.1158/0008-5472.CAN-22-1535. Cancer Res. 2022. PMID: 35819255 Free PMC article. Review.
Cited by
- Functional Validation of Rare Human Genetic Variants Involved in Homologous Recombination Using Saccharomyces cerevisiae.
Lee MS, Yu M, Kim KY, Park GH, Kwack K, Kim KP. Lee MS, et al. PLoS One. 2015 May 4;10(5):e0124152. doi: 10.1371/journal.pone.0124152. eCollection 2015. PLoS One. 2015. PMID: 25938495 Free PMC article. - RIF1 counteracts BRCA1-mediated end resection during DNA repair.
Feng L, Fong KW, Wang J, Wang W, Chen J. Feng L, et al. J Biol Chem. 2013 Apr 19;288(16):11135-43. doi: 10.1074/jbc.M113.457440. Epub 2013 Mar 13. J Biol Chem. 2013. PMID: 23486525 Free PMC article. - Complementation of hypersensitivity to DNA interstrand crosslinking agents demonstrates that XRCC2 is a Fanconi anaemia gene.
Park JY, Virts EL, Jankowska A, Wiek C, Othman M, Chakraborty SC, Vance GH, Alkuraya FS, Hanenberg H, Andreassen PR. Park JY, et al. J Med Genet. 2016 Oct;53(10):672-680. doi: 10.1136/jmedgenet-2016-103847. Epub 2016 May 20. J Med Genet. 2016. PMID: 27208205 Free PMC article. - Star wars against leukemia: attacking the clones.
Toma MM, Skorski T. Toma MM, et al. Leukemia. 2024 Nov;38(11):2293-2302. doi: 10.1038/s41375-024-02369-6. Epub 2024 Sep 2. Leukemia. 2024. PMID: 39223295 Free PMC article. Review. - PALB2 (partner and localizer of BRCA2).
Hanenberg H, Andreassen PR. Hanenberg H, et al. Atlas Genet Cytogenet Oncol Haematol. 2018 Apr;22(12):484-490. doi: 10.4267/2042/69016. Atlas Genet Cytogenet Oncol Haematol. 2018. PMID: 31413733 Free PMC article.
References
- Jemal A, et al. Cancer Statistics, 2009. CA Cancer J Clin. 2009 - PubMed
- Gudmundsdottir K, Ashworth A. The roles of BRCA1 and BRCA2 and associated proteins in the maintenance of genomic stability. Oncogene. 2006;25:5864–5874. - PubMed
- Venkitaraman AR. Linking the cellular functions of BRCA genes to cancer pathogenesis and treatment. Annu Rev Pathol. 2009;4:461–487. - PubMed
- West SC. Molecular views of recombination proteins and their control. Nat Rev Mol Cell Biol. 2003;4:435–445. - PubMed
- Davies AA, et al. Role of BRCA2 in control of the RAD51 recombination and DNA repair protein. Mol Cell. 2001;7:273–282. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 CA138804/CA/NCI NIH HHS/United States
- R01 CA138804-03/CA/NCI NIH HHS/United States
- R01CA138804/CA/NCI NIH HHS/United States
- R01 CA138804-01A1/CA/NCI NIH HHS/United States
- R01 CA138804-02/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous