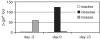Single-strand nicks induce homologous recombination with less toxicity than double-strand breaks using an AAV vector template - PubMed (original) (raw)
Single-strand nicks induce homologous recombination with less toxicity than double-strand breaks using an AAV vector template
Michael J Metzger et al. Nucleic Acids Res. 2011 Feb.
Abstract
Gene targeting by homologous recombination (HR) can be induced by double-strand breaks (DSBs), however these breaks can be toxic and potentially mutagenic. We investigated the I-AniI homing endonuclease engineered to produce only nicks, and found that nicks induce HR with both plasmid and adeno-associated virus (AAV) vector templates. The rates of nick-induced HR were lower than with DSBs (24-fold lower for plasmid transfection and 4- to 6-fold lower for AAV vector infection), but they still represented a significant increase over background (240- and 30-fold, respectively). We observed severe toxicity with the I-AniI 'cleavase', but no evidence of toxicity with the I-AniI 'nickase.' Additionally, the frequency of nickase-induced mutations at the I-AniI site was at least 150-fold lower than that induced by the cleavase. These results, and the observation that the surrounding sequence context of a target site affects nick-induced HR but not DSB-induced HR, strongly argue that nicks induce HR through a different mechanism than DSBs, allowing for gene correction without the toxicity and mutagenic activity of DSBs.
Figures
Figure 1.
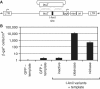
Nicks induce HR with a transfected template. (A) Schematic of integrated target site CnZPNOA3, with a lacZ gene inactivated by the introduction of the I-AniI recognition site (black box). The repair template plasmid (pA2nZ3113) is shown above with regions of homology indicated by a dotted ‘X’. (B) β-gal+ cells per cm2 in confluent polyclonal 293/CnZPNOA3 cells transfected with one of the plasmids expressing either GFP or the inactive, cleavase or nickase I-AniI and template plasmid pA2nZ3113 (except for the GFP−template condition, in which cells were transfected with only GFP-expressing plasmid). Values are means of four experiments ± SE.
Figure 2.
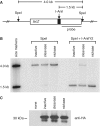
Expression of I-AniI cleavase generates DSBs, but nickase does not. (A) Schematic of the integrated target site CnZPNOA3, containing the I-AniI recognition site, with probe and enzyme cleavage sites marked. (B) Genomic DNA was extracted from 293/CnZPNOA3 cells 3 days after transduction with I-AniI-expressing lentivirus—either inactive, cleavase or nickase. DNA was digested in vitro with either SpeI alone or SpeI and purified I-AniI cleavase before being used for the Southern blot. Size markers were generated by in vitro digestion of pCnZPNOA2 plasmid with SpeI and SpeI/I-AniI and gel extraction of 4.0- and 1.5-kb bands, respectively. (C) Cell lysate was collected from 293/CnZPNOA3 cells 3 days after transduction with the same I-AniI-expressing lentivirus preparations used for the Southern blot and further HR assays. An amount of 10 µg of each lysate was loaded for western analysis and anti-HA antibody (mouse mAb 6E2, Cell Signaling Technology) was used to detect the HA-tagged I-AniI proteins.
Figure 3.
Analysis of potential NHEJ mutations using a sensitive qPCR-based assay. (A) Schematic of the integrated target site CnZPNOA3 showing the positions of the primers used for qPCR: lacZqF1 (F1), lacZqR1 (R1), lacZqF2 (F2) and lacZqR2 (R2). (B) Table showing the ratio of qPCR amplification with primer pair 2 to primer pair 1 after digest of genomic DNA with I-AniI, analysis of the fraction of the qPCR products which cannot be cleaved by I-AniI, and the final estimated percentage of mutated I-AniI sites (a product of the first two columns). Values are means of two experiments calculated independently (individual values for nickase-induced mutation were 120- and 180-fold less than that with cleavase). (C) Primer pair 2 qPCR products were digested with I-AniI, and a representative agarose gel revealing the digested and undigested products is shown.
Figure 4.
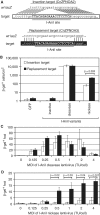
Nicks induce HR with an AAV vector template with less toxicity than DSBs. (A) Schematic of integrated target site CnZPNOA3, with a lacZ gene inactivated by the introduction of the I-AniI recognition site. The AAV2-nZ3113 genome used as an HR template is shown above with homology indicated by a dotted ‘X’. (B) β-gal+ foci counted 9 days after infection of 5 × 104 cells with 1.5 × 104 vg/cell AAV2-nZ3113. Cells were transduced with different MOIs of the three I-AniI-expressing lentivirus vectors 3 days prior to AAV vector delivery. (C) Survival of cells transduced with different MOIs of the three I-AniI-expressing lentivirus vectors determined by counting Coomassie-stained colonies 9 days after AAV vector infection and normalized to colony-forming ability of untransduced cells assayed in parallel. Survival points and β-gal+ bars represent means of three experiments ± SE, each conducted with independent polyclonal cell populations. White bars and white squares represent inactive I-AniI-expressing lentivirus; black bars and black diamonds, I-AniI cleavase; gray bars and gray circles, I-AniI nickase. (D) β-gal+ foci counted 9 days after infection of 5 × 104 cells with varying amounts of AAV2-nZ3113. All cells were transduced with I-AniI-expressing lentivirus vectors 3 days earlier at an MOI of 1 TU/cell. White bars represent mock-lentiviral infection; black bars, cleavase; gray bars, nickase.
Figure 5.
Timing of endonuclease delivery. β-gal+ foci counted 9 days after infection of 293/CnZPNOA3 cells with 2 × 104 vg/cell of AAV2-nZ3113. In all cases, cells were seeded at 5 × 104 cells per well in 24-well plates the day before AAV vector infection. Cells were transduced with one of three I-AniI-expressing lentivirus vectors (at an MOI of 2.5 TU/cell) at three different times: 3 days before, the same time as, or 3 days after AAV vector infection. Results are means of two experiments, except for the transduction 3 days after AAV vector infection and the nickase transduction at the same time as AAV vector infection, which are means of three experiments. White bars represent inactive I-AniI-expressing lentivirus; black bars, I-AniI cleavase; gray bars, I-AniI nickase.
Figure 6.
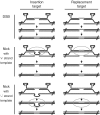
Target design affects nick-induced HR, but not DSB-induced HR. (A) Schematic of the differences between the insertion (CnZPNOA2) and replacement (CnZPNOA3) targets with homology to wt lacZ. The I-AniI recognition site is capitalized and in bold for both targets. (B) β-gal+ cells/cm2 after transfection as in Figure 2, using 293 cells with integrated insertion target (CnZPNOA2, white bars) or replacement target (CnZPNOA3, black bars). Means of four experiments ± SE. (C) β-gal+ foci after transduction with cleavase-expressing lentivirus and infection with AAV2-nZ3113 as in Figure 3. Means of three experiments ± SE. (D) β-gal+ foci after transduction with nickase-expressing lentivirus and infection with AAV2-nZ3113 as in Figure 3. Means of three experiments ± SE.
Figure 7.
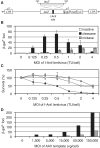
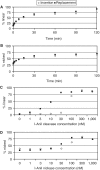
In vitro comparison of replacement and insertion targets. (A) Frequency of linearized plasmid containing either the insertion target (pnZA2, white diamonds) or replacement (pnZA3, black squares) following in vitro digestion with 1 µM purified I-AniI cleavase at 37°C for 0–2 h. (B) Frequency of nicked plasmids with 1 µM purified I-AniI nickase. (C) Frequency of linearized plasmid with both targets with increasing concentrations of purified I-AniI cleavase for 2 h at 37°C in vitro. (D) Frequency of nicked plasmid with increasing concentrations of purified I-AniI nickase for 2 h at 37°C in vitro.
Figure 8.
Model for HR at DSBs and nicks. When either the replacement or insertion target sites are cleaved, the ends will be processed and the AAV vector template will invade and align with one of the broken strands. When the replacement and insertion sites are nicked, however, the alignment of the target and AAV vector template is different. The nicked target can align with a single-stranded AAV vector template of either ‘+’ or ‘–’ orientation, followed by Holiday junction resolution and completion of HR. Notably, in nick-induced HR with either template there is a step in which an intact heterodimer is present (marked with a gray circle). This does not occur if both strands are cut. We hypothesize that the discordant activity observed in Figure 6 could be explained by discrimination at the level of resolution of this heterodimer.
Similar articles
- PARP-mediated repair, homologous recombination, and back-up non-homologous end joining-like repair of single-strand nicks.
Metzger MJ, Stoddard BL, Monnat RJ Jr. Metzger MJ, et al. DNA Repair (Amst). 2013 Jul;12(7):529-34. doi: 10.1016/j.dnarep.2013.04.004. Epub 2013 May 16. DNA Repair (Amst). 2013. PMID: 23684799 Free PMC article. - Self-complementary AAV mediates gene targeting and enhances endonuclease delivery for double-strand break repair.
Hirsch ML, Green L, Porteus MH, Samulski RJ. Hirsch ML, et al. Gene Ther. 2010 Sep;17(9):1175-80. doi: 10.1038/gt.2010.65. Epub 2010 May 13. Gene Ther. 2010. PMID: 20463753 Free PMC article. - Fate of recombinant adeno-associated viral vector genomes during DNA double-strand break-induced gene targeting in human cells.
Gellhaus K, Cornu TI, Heilbronn R, Cathomen T. Gellhaus K, et al. Hum Gene Ther. 2010 May;21(5):543-53. doi: 10.1089/hum.2009.167. Hum Gene Ther. 2010. PMID: 20021219 - Nick-initiated homologous recombination: Protecting the genome, one strand at a time.
Vriend LE, Krawczyk PM. Vriend LE, et al. DNA Repair (Amst). 2017 Feb;50:1-13. doi: 10.1016/j.dnarep.2016.12.005. Epub 2016 Dec 29. DNA Repair (Amst). 2017. PMID: 28087249 Review. - Initiation of homologous recombination at DNA nicks.
Maizels N, Davis L. Maizels N, et al. Nucleic Acids Res. 2018 Aug 21;46(14):6962-6973. doi: 10.1093/nar/gky588. Nucleic Acids Res. 2018. PMID: 29986051 Free PMC article. Review.
Cited by
- Concerted nicking of donor and chromosomal acceptor DNA promotes homology-directed gene targeting in human cells.
Gonçalves MA, van Nierop GP, Holkers M, de Vries AA. Gonçalves MA, et al. Nucleic Acids Res. 2012 Apr;40(8):3443-55. doi: 10.1093/nar/gkr1234. Epub 2011 Dec 20. Nucleic Acids Res. 2012. PMID: 22189101 Free PMC article. - Efficient CRISPR-Cas9-mediated generation of knockin human pluripotent stem cells lacking undesired mutations at the targeted locus.
Merkle FT, Neuhausser WM, Santos D, Valen E, Gagnon JA, Maas K, Sandoe J, Schier AF, Eggan K. Merkle FT, et al. Cell Rep. 2015 May 12;11(6):875-883. doi: 10.1016/j.celrep.2015.04.007. Epub 2015 Apr 30. Cell Rep. 2015. PMID: 25937281 Free PMC article. - The democratization of gene editing: Insights from site-specific cleavage and double-strand break repair.
Jasin M, Haber JE. Jasin M, et al. DNA Repair (Amst). 2016 Aug;44:6-16. doi: 10.1016/j.dnarep.2016.05.001. Epub 2016 May 12. DNA Repair (Amst). 2016. PMID: 27261202 Free PMC article. Review. - DNA double-strand breaks as a method of radiation measurements for therapeutic beams.
Obeidat M, McConnell KA, Li X, Bui B, Stathakis S, Papanikolaou N, Rasmussen K, Ha CS, Lee SE, Shim EY, Kirby N. Obeidat M, et al. Med Phys. 2018 Jul;45(7):3460-3465. doi: 10.1002/mp.12956. Epub 2018 May 23. Med Phys. 2018. PMID: 29745994 Free PMC article. - Inducing multiple nicks promotes interhomolog homologous recombination to correct heterozygous mutations in somatic cells.
Tomita A, Sasanuma H, Owa T, Nakazawa Y, Shimada M, Fukuoka T, Ogi T, Nakada S. Tomita A, et al. Nat Commun. 2023 Sep 15;14(1):5607. doi: 10.1038/s41467-023-41048-5. Nat Commun. 2023. PMID: 37714828 Free PMC article.
References
- Miller DG, Wang PR, Petek LM, Hirata RK, Sands MS, Russell DW. Gene targeting in vivo by adeno-associated virus vectors. Nat. Biotechnol. 2006;24:1022–1026. - PubMed
- Urnov FD, Miller JC, Lee YL, Beausejour CM, Rock JM, Augustus S, Jamieson AC, Porteus MH, Gregory PD, Holmes MC. Highly efficient endogenous human gene correction using designed zinc-finger nucleases. Nature. 2005;435:646–651. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources