Structural basis of carbohydrate recognition by calreticulin - PubMed (original) (raw)
Structural basis of carbohydrate recognition by calreticulin
Guennadi Kozlov et al. J Biol Chem. 2010.
Abstract
The calnexin cycle is a process by which glycosylated proteins are subjected to folding cycles in the endoplasmic reticulum lumen via binding to the membrane protein calnexin (CNX) or to its soluble homolog calreticulin (CRT). CNX and CRT specifically recognize monoglucosylated Glc(1)Man(9)GlcNAc(2) glycans, but the structural determinants underlying this specificity are unknown. Here, we report a 1.95-Å crystal structure of the CRT lectin domain in complex with the tetrasaccharide α-Glc-(1→3)-α-Man-(1→2)-α-Man-(1→2)-Man. The tetrasaccharide binds to a long channel on CRT formed by a concave β-sheet. All four sugar moieties are engaged in the protein binding via an extensive network of hydrogen bonds and hydrophobic contacts. The structure explains the requirement for glucose at the nonreducing end of the carbohydrate; the oxygen O(2) of glucose perfectly fits to a pocket formed by CRT side chains while forming direct hydrogen bonds with the carbonyl of Gly(124) and the side chain of Lys(111). The structure also explains a requirement for the Cys(105)-Cys(137) disulfide bond in CRT/CNX for efficient carbohydrate binding. The Cys(105)-Cys(137) disulfide bond is involved in intimate contacts with the third and fourth sugar moieties of the Glc(1)Man(3) tetrasaccharide. Finally, the structure rationalizes previous mutagenesis of CRT and lays a structural groundwork for future studies of the role of CNX/CRT in diverse biological pathways.
Figures
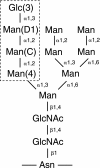
SCHEME 1.
Structure of monoglucosylated _N_-linked glycan. The labels 3, D1, C, and 4 indicate the sugar positions. The tetrasaccharide that binds CRT is boxed.
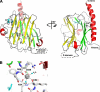
FIGURE 1.
Structure of the CRT lectin domain. A, schematic representation of the structure. Helices are shown in red, β-strands in the concave β-sheet are yellow, the convex β-sheet is green, and the two additional strands β2 and β3 are cyan. The bound calcium ion is shown as a gray sphere. The position of the P-domain that contains strands β13–β20 is indicated by a dashed line. Cys105 and Cys137 in the concave β-sheet form a disulfide bond (S–S). B, enlarged view of the calcium-binding site. Residues and hydrogen bonds (dashed lines) coordinating the calcium ion are shown along with two coordinating water molecules (cyan spheres). Residue color coding is the same as in A. N-term, N terminus; C-term, C terminus.
FIGURE 2.
Structural basis of Glc1Man3 recognition by CRT. A, omit map calculated in the absence of tetrasaccharide shows well defined electron density (blue) for all four sugar moieties. The tetrasaccharide binds in a cavity on the concave β-sheet. B, surface representation of CRT shows the side chains of Phe74, Met131, His145, Ile147, Trp319, and the Cys105–Cys137 disulfide bridge form the walls of the cavity in contact with the glycan (magenta). C, oxygens in the tetrasaccharide form a network of hydrogen bonds (dotted lines) with ordered water molecules (cyan spheres) and CRT. Residues that disrupt CRT binding when mutated are shown in gray (17–19). D, the equatorial oxygen (O2) of glucose makes hydrogen bonds with the side chain of Lys111 and backbone carbonyl of Gly124. Mannose has an axial O2, which clashes sterically with the underlying side chain of Met131 to prevent binding in that position.
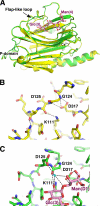
FIGURE 3.
CRT undergoes limited conformational changes upon carbohydrate binding. A, overlay of unliganded (yellow) and Glc1Man3-bound (green) CRT lectin domain structures shows differences in the large loop between strands β6 and β7. B, the conformation of the loop in unliganded CRT is stabilized by hydrogen bonds between the side chain of Asp317 and amides of Gly124 and Asp125 and between the side chain of Lys111 and carbonyl of Asp125. C, in the complex, sugar residues Glc(3) and Man(D1) engage loop residues Gly124 and Asp125 through a 60 ° rotation in the ψ backbone angle of Gly124. This rotation enables the side chain of Asp125 to participate in carbohydrate binding via an ordered water molecule (cyan sphere). The interactions of Asp317 with Gly124 are replaced by hydrogen bonds of Asp317 with Man(D1).
FIGURE 4.
Structural comparison of the CRT and CNX lectin domains. A, overlay of the CRT (yellow) and CNX (Protein Data Bank code 1JHN; cyan) structures. The termini and boundaries of the P-domain of CRT are indicated. B, structure-based sequence alignment of mouse CRT and dog CNX lectin domains. Secondary structure of CRT is shown above the alignment with β-strands and α-helices labeled. Residues that make direct hydrogen bonds with Glc1Man3 are highlighted in cyan, and those making van der Waals contacts are highlighted in gray. The position of the internal P-domain comprising strands β13 to β18 of CRT is indicated. C, enlarged view of the carbohydrate-binding site shows nearly identical positioning of key residues in CRT and CNX. Residue numbers refer to CRT. N-term, N terminus; C-term, C terminus.
FIGURE 5.
Structural model of full-length CRT. The lectin domain is shown in green, and the bound carbohydrate is shown in magenta. The approximate orientation of the P-domain (dark blue) is shown based on the CNX structure with one repeat unit removed to match the shorter CRT P-domain. The bound carbohydrate is shown as part of the _N_-linked glycan linked to an asparagine residue of an unfolded protein. The C terminus of CRT contains a Glu-, Asp-rich sequence, which binds calcium, and a KDEL ER retention signal. The residues that define the boundaries of crystallized CRT fragment and the portion of the P-domain that binds the thiol oxidoreductase, ERp57, are labeled.
Similar articles
- Role of N-oligosaccharide endoplasmic reticulum processing reactions in glycoprotein folding and degradation.
Parodi AJ. Parodi AJ. Biochem J. 2000 May 15;348 Pt 1(Pt 1):1-13. Biochem J. 2000. PMID: 10794707 Free PMC article. Review. - Mutational analysis provides molecular insight into the carbohydrate-binding region of calreticulin: pivotal roles of tyrosine-109 and aspartate-135 in carbohydrate recognition.
Kapoor M, Ellgaard L, Gopalakrishnapai J, Schirra C, Gemma E, Oscarson S, Helenius A, Surolia A. Kapoor M, et al. Biochemistry. 2004 Jan 13;43(1):97-106. doi: 10.1021/bi0355286. Biochemistry. 2004. PMID: 14705935 - Delineation of the lectin site of the molecular chaperone calreticulin.
Thomson SP, Williams DB. Thomson SP, et al. Cell Stress Chaperones. 2005 Autumn;10(3):242-51. doi: 10.1379/csc-126.1. Cell Stress Chaperones. 2005. PMID: 16184769 Free PMC article. - Calnexin/Calreticulin and Assays Related to N-Glycoprotein Folding In Vitro.
Ihara Y, Ikezaki M, Takatani M, Ito Y. Ihara Y, et al. Methods Mol Biol. 2020;2132:295-308. doi: 10.1007/978-1-0716-0430-4_29. Methods Mol Biol. 2020. PMID: 32306337 - Protein glucosylation and its role in protein folding.
Parodi AJ. Parodi AJ. Annu Rev Biochem. 2000;69:69-93. doi: 10.1146/annurev.biochem.69.1.69. Annu Rev Biochem. 2000. PMID: 10966453 Review.
Cited by
- The Role of Molecular Flexibility in Antigen Presentation and T Cell Receptor-Mediated Signaling.
Natarajan K, Jiang J, May NA, Mage MG, Boyd LF, McShan AC, Sgourakis NG, Bax A, Margulies DH. Natarajan K, et al. Front Immunol. 2018 Jul 17;9:1657. doi: 10.3389/fimmu.2018.01657. eCollection 2018. Front Immunol. 2018. PMID: 30065727 Free PMC article. Review. - Structural basis for two-step glucose trimming by glucosidase II involved in ER glycoprotein quality control.
Satoh T, Toshimori T, Yan G, Yamaguchi T, Kato K. Satoh T, et al. Sci Rep. 2016 Feb 5;6:20575. doi: 10.1038/srep20575. Sci Rep. 2016. PMID: 26847925 Free PMC article. - Considerations for Glycoprotein Production.
Clarke EC. Clarke EC. Methods Mol Biol. 2024;2762:329-351. doi: 10.1007/978-1-0716-3666-4_20. Methods Mol Biol. 2024. PMID: 38315375 - Emerging structural insights into glycoprotein quality control coupled with N-glycan processing in the endoplasmic reticulum.
Satoh T, Yamaguchi T, Kato K. Satoh T, et al. Molecules. 2015 Jan 30;20(2):2475-91. doi: 10.3390/molecules20022475. Molecules. 2015. PMID: 25647580 Free PMC article. Review. - Protein secretion and the endoplasmic reticulum.
Benham AM. Benham AM. Cold Spring Harb Perspect Biol. 2012 Aug 1;4(8):a012872. doi: 10.1101/cshperspect.a012872. Cold Spring Harb Perspect Biol. 2012. PMID: 22700933 Free PMC article. Review.
References
- Helenius A., Aebi M. (2004) Annu. Rev. Biochem. 73, 1019–1049 - PubMed
- Lederkremer G. Z. (2009) Curr. Opin. Struct. Biol. 19, 515–523 - PubMed
- Ware F. E., Vassilakos A., Peterson P. A., Jackson M. R., Lehrman M. A., Williams D. B. (1995) J. Biol. Chem. 270, 4697–4704 - PubMed
- Spiro R. G., Zhu Q., Bhoyroo V., Söling H. D. (1996) J. Biol. Chem. 271, 11588–11594 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- MOP-81277/CAPMC/ CIHR/Canada
- P41 RR001646/RR/NCRR NIH HHS/United States
- RR-01646/RR/NCRR NIH HHS/United States
- MOP-53310/CAPMC/ CIHR/Canada
- DMR0225180/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials