Hundreds of variants clustered in genomic loci and biological pathways affect human height - PubMed (original) (raw)
. 2010 Oct 14;467(7317):832-8.
doi: 10.1038/nature09410. Epub 2010 Sep 29.
Karol Estrada, Guillaume Lettre, Sonja I Berndt, Michael N Weedon, Fernando Rivadeneira, Cristen J Willer, Anne U Jackson, Sailaja Vedantam, Soumya Raychaudhuri, Teresa Ferreira, Andrew R Wood, Robert J Weyant, Ayellet V Segrè, Elizabeth K Speliotes, Eleanor Wheeler, Nicole Soranzo, Ju-Hyun Park, Jian Yang, Daniel Gudbjartsson, Nancy L Heard-Costa, Joshua C Randall, Lu Qi, Albert Vernon Smith, Reedik Mägi, Tomi Pastinen, Liming Liang, Iris M Heid, Jian'an Luan, Gudmar Thorleifsson, Thomas W Winkler, Michael E Goddard, Ken Sin Lo, Cameron Palmer, Tsegaselassie Workalemahu, Yurii S Aulchenko, Asa Johansson, M Carola Zillikens, Mary F Feitosa, Tõnu Esko, Toby Johnson, Shamika Ketkar, Peter Kraft, Massimo Mangino, Inga Prokopenko, Devin Absher, Eva Albrecht, Florian Ernst, Nicole L Glazer, Caroline Hayward, Jouke-Jan Hottenga, Kevin B Jacobs, Joshua W Knowles, Zoltán Kutalik, Keri L Monda, Ozren Polasek, Michael Preuss, Nigel W Rayner, Neil R Robertson, Valgerdur Steinthorsdottir, Jonathan P Tyrer, Benjamin F Voight, Fredrik Wiklund, Jianfeng Xu, Jing Hua Zhao, Dale R Nyholt, Niina Pellikka, Markus Perola, John R B Perry, Ida Surakka, Mari-Liis Tammesoo, Elizabeth L Altmaier, Najaf Amin, Thor Aspelund, Tushar Bhangale, Gabrielle Boucher, Daniel I Chasman, Constance Chen, Lachlan Coin, Matthew N Cooper, Anna L Dixon, Quince Gibson, Elin Grundberg, Ke Hao, M Juhani Junttila, Lee M Kaplan, Johannes Kettunen, Inke R König, Tony Kwan, Robert W Lawrence, Douglas F Levinson, Mattias Lorentzon, Barbara McKnight, Andrew P Morris, Martina Müller, Julius Suh Ngwa, Shaun Purcell, Suzanne Rafelt, Rany M Salem, Erika Salvi, Serena Sanna, Jianxin Shi, Ulla Sovio, John R Thompson, Michael C Turchin, Liesbeth Vandenput, Dominique J Verlaan, Veronique Vitart, Charles C White, Andreas Ziegler, Peter Almgren, Anthony J Balmforth, Harry Campbell, Lorena Citterio, Alessandro De Grandi, Anna Dominiczak, Jubao Duan, Paul Elliott, Roberto Elosua, Johan G Eriksson, Nelson B Freimer, Eco J C Geus, Nicola Glorioso, Shen Haiqing, Anna-Liisa Hartikainen, Aki S Havulinna, Andrew A Hicks, Jennie Hui, Wilmar Igl, Thomas Illig, Antti Jula, Eero Kajantie, Tuomas O Kilpeläinen, Markku Koiranen, Ivana Kolcic, Seppo Koskinen, Peter Kovacs, Jaana Laitinen, Jianjun Liu, Marja-Liisa Lokki, Ana Marusic, Andrea Maschio, Thomas Meitinger, Antonella Mulas, Guillaume Paré, Alex N Parker, John F Peden, Astrid Petersmann, Irene Pichler, Kirsi H Pietiläinen, Anneli Pouta, Martin Ridderstråle, Jerome I Rotter, Jennifer G Sambrook, Alan R Sanders, Carsten Oliver Schmidt, Juha Sinisalo, Jan H Smit, Heather M Stringham, G Bragi Walters, Elisabeth Widen, Sarah H Wild, Gonneke Willemsen, Laura Zagato, Lina Zgaga, Paavo Zitting, Helene Alavere, Martin Farrall, Wendy L McArdle, Mari Nelis, Marjolein J Peters, Samuli Ripatti, Joyce B J van Meurs, Katja K Aben, Kristin G Ardlie, Jacques S Beckmann, John P Beilby, Richard N Bergman, Sven Bergmann, Francis S Collins, Daniele Cusi, Martin den Heijer, Gudny Eiriksdottir, Pablo V Gejman, Alistair S Hall, Anders Hamsten, Heikki V Huikuri, Carlos Iribarren, Mika Kähönen, Jaakko Kaprio, Sekar Kathiresan, Lambertus Kiemeney, Thomas Kocher, Lenore J Launer, Terho Lehtimäki, Olle Melander, Tom H Mosley Jr, Arthur W Musk, Markku S Nieminen, Christopher J O'Donnell, Claes Ohlsson, Ben Oostra, Lyle J Palmer, Olli Raitakari, Paul M Ridker, John D Rioux, Aila Rissanen, Carlo Rivolta, Heribert Schunkert, Alan R Shuldiner, David S Siscovick, Michael Stumvoll, Anke Tönjes, Jaakko Tuomilehto, Gert-Jan van Ommen, Jorma Viikari, Andrew C Heath, Nicholas G Martin, Grant W Montgomery, Michael A Province, Manfred Kayser, Alice M Arnold, Larry D Atwood, Eric Boerwinkle, Stephen J Chanock, Panos Deloukas, Christian Gieger, Henrik Grönberg, Per Hall, Andrew T Hattersley, Christian Hengstenberg, Wolfgang Hoffman, G Mark Lathrop, Veikko Salomaa, Stefan Schreiber, Manuela Uda, Dawn Waterworth, Alan F Wright, Themistocles L Assimes, Inês Barroso, Albert Hofman, Karen L Mohlke, Dorret I Boomsma, Mark J Caulfield, L Adrienne Cupples, Jeanette Erdmann, Caroline S Fox, Vilmundur Gudnason, Ulf Gyllensten, Tamara B Harris, Richard B Hayes, Marjo-Riitta Jarvelin, Vincent Mooser, Patricia B Munroe, Willem H Ouwehand, Brenda W Penninx, Peter P Pramstaller, Thomas Quertermous, Igor Rudan, Nilesh J Samani, Timothy D Spector, Henry Völzke, Hugh Watkins, James F Wilson, Leif C Groop, Talin Haritunians, Frank B Hu, Robert C Kaplan, Andres Metspalu, Kari E North, David Schlessinger, Nicholas J Wareham, David J Hunter, Jeffrey R O'Connell, David P Strachan, H-Erich Wichmann, Ingrid B Borecki, Cornelia M van Duijn, Eric E Schadt, Unnur Thorsteinsdottir, Leena Peltonen, André G Uitterlinden, Peter M Visscher, Nilanjan Chatterjee, Ruth J F Loos, Michael Boehnke, Mark I McCarthy, Erik Ingelsson, Cecilia M Lindgren, Gonçalo R Abecasis, Kari Stefansson, Timothy M Frayling, Joel N Hirschhorn
Affiliations
- PMID: 20881960
- PMCID: PMC2955183
- DOI: 10.1038/nature09410
Hundreds of variants clustered in genomic loci and biological pathways affect human height
Hana Lango Allen et al. Nature. 2010.
Abstract
Most common human traits and diseases have a polygenic pattern of inheritance: DNA sequence variants at many genetic loci influence the phenotype. Genome-wide association (GWA) studies have identified more than 600 variants associated with human traits, but these typically explain small fractions of phenotypic variation, raising questions about the use of further studies. Here, using 183,727 individuals, we show that hundreds of genetic variants, in at least 180 loci, influence adult height, a highly heritable and classic polygenic trait. The large number of loci reveals patterns with important implications for genetic studies of common human diseases and traits. First, the 180 loci are not random, but instead are enriched for genes that are connected in biological pathways (P = 0.016) and that underlie skeletal growth defects (P < 0.001). Second, the likely causal gene is often located near the most strongly associated variant: in 13 of 21 loci containing a known skeletal growth gene, that gene was closest to the associated variant. Third, at least 19 loci have multiple independently associated variants, suggesting that allelic heterogeneity is a frequent feature of polygenic traits, that comprehensive explorations of already-discovered loci should discover additional variants and that an appreciable fraction of associated loci may have been identified. Fourth, associated variants are enriched for likely functional effects on genes, being over-represented among variants that alter amino-acid structure of proteins and expression levels of nearby genes. Our data explain approximately 10% of the phenotypic variation in height, and we estimate that unidentified common variants of similar effect sizes would increase this figure to approximately 16% of phenotypic variation (approximately 20% of heritable variation). Although additional approaches are needed to dissect the genetic architecture of polygenic human traits fully, our findings indicate that GWA studies can identify large numbers of loci that implicate biologically relevant genes and pathways.
Figures
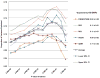
Figure 1. Phenotypic variance explained by common variants
(a) Variance explained is higher when SNPs not reaching genome-wide significance are included in the prediction model. The y-axis represents the proportion of variance explained at different _P_-value thresholds from Stage 1. Results are given for five studies that were not part of Stage 1. *Proportion of variation explained by the 180 SNPs. (b) Cumulative number of susceptibility loci expected to be discovered, including already identified loci and as yet undetected loci. The projections are based on loci that achieved a significance level of P<5×10-8 in the initial scan and the distribution of their effect sizes in Stage 2. The dotted red line corresponds to expected phenotypic variance explained by the 110 loci that reached genome-wide significance in Stage 1, were replicated in Stage 2 and had at least 1% power.
Figure 1. Phenotypic variance explained by common variants
(a) Variance explained is higher when SNPs not reaching genome-wide significance are included in the prediction model. The y-axis represents the proportion of variance explained at different _P_-value thresholds from Stage 1. Results are given for five studies that were not part of Stage 1. *Proportion of variation explained by the 180 SNPs. (b) Cumulative number of susceptibility loci expected to be discovered, including already identified loci and as yet undetected loci. The projections are based on loci that achieved a significance level of P<5×10-8 in the initial scan and the distribution of their effect sizes in Stage 2. The dotted red line corresponds to expected phenotypic variance explained by the 110 loci that reached genome-wide significance in Stage 1, were replicated in Stage 2 and had at least 1% power.
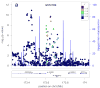
Figure 2. Example of a locus with a secondary signal before (a) and after (b) conditioning
The plot is centered on the conditioned SNP (purple diamond) at the locus. r2 is based on the CEU HapMap II samples. The blue line and right hand Y axis represent CEU HapMap II recombination rates. Created using LocusZoom (
http://csg.sph.umich.edu/locuszoom/
).
Figure 2. Example of a locus with a secondary signal before (a) and after (b) conditioning
The plot is centered on the conditioned SNP (purple diamond) at the locus. r2 is based on the CEU HapMap II samples. The blue line and right hand Y axis represent CEU HapMap II recombination rates. Created using LocusZoom (
http://csg.sph.umich.edu/locuszoom/
).
Figure 3. Loci associated with height contain genes related to each other
(a) 180 height-associated SNPs. The y-axis plots GRAIL _P_-values on a log scale. The histogram corresponds to the distribution of GRAIL _P_-values for 1,000 sets of 180 matched SNPs. The scatter plot represents GRAIL results for the 180 height SNPs (blue dots). The black horizontal line marks the median of the GRAIL _P_-values (_P_=0.14). The top 10 keywords linking the genes were: ‘growth’, ‘kinase’, ‘factor’, ‘transcription’, ‘signaling’, ‘binding’, ‘differentiation’, ‘development’, ‘insulin’, ‘bone’. (b) Graphical representation of the connections between SNPs and corresponding genes for the 42 SNPs with GRAIL P<0.01. Thicker and redder lines imply stronger literature-based connectivity.
Similar articles
- Genome-wide Analysis of Body Proportion Classifies Height-Associated Variants by Mechanism of Action and Implicates Genes Important for Skeletal Development.
Chan Y, Salem RM, Hsu YH, McMahon G, Pers TH, Vedantam S, Esko T, Guo MH, Lim ET; GIANT Consortium; Franke L, Smith GD, Strachan DP, Hirschhorn JN. Chan Y, et al. Am J Hum Genet. 2015 May 7;96(5):695-708. doi: 10.1016/j.ajhg.2015.02.018. Epub 2015 Apr 9. Am J Hum Genet. 2015. PMID: 25865494 Free PMC article. - A Selection Operator for Summary Association Statistics Reveals Allelic Heterogeneity of Complex Traits.
Ning Z, Lee Y, Joshi PK, Wilson JF, Pawitan Y, Shen X. Ning Z, et al. Am J Hum Genet. 2017 Dec 7;101(6):903-912. doi: 10.1016/j.ajhg.2017.09.027. Am J Hum Genet. 2017. PMID: 29198721 Free PMC article. - Common variants show predicted polygenic effects on height in the tails of the distribution, except in extremely short individuals.
Chan Y, Holmen OL, Dauber A, Vatten L, Havulinna AS, Skorpen F, Kvaløy K, Silander K, Nguyen TT, Willer C, Boehnke M, Perola M, Palotie A, Salomaa V, Hveem K, Frayling TM, Hirschhorn JN, Weedon MN. Chan Y, et al. PLoS Genet. 2011 Dec;7(12):e1002439. doi: 10.1371/journal.pgen.1002439. Epub 2011 Dec 29. PLoS Genet. 2011. PMID: 22242009 Free PMC article. - Genome-wide association studies of human growth traits.
Weedon MN. Weedon MN. Nestle Nutr Inst Workshop Ser. 2013;71:29-38. doi: 10.1159/000342541. Epub 2013 Jan 22. Nestle Nutr Inst Workshop Ser. 2013. PMID: 23502136 Review. - Research review: Polygenic methods and their application to psychiatric traits.
Wray NR, Lee SH, Mehta D, Vinkhuyzen AA, Dudbridge F, Middeldorp CM. Wray NR, et al. J Child Psychol Psychiatry. 2014 Oct;55(10):1068-87. doi: 10.1111/jcpp.12295. Epub 2014 Aug 1. J Child Psychol Psychiatry. 2014. PMID: 25132410 Review.
Cited by
- Analyses of GWAS signal using GRIN identify additional genes contributing to suicidal behavior.
Sullivan KA, Lane M, Cashman M, Miller JI, Pavicic M, Walker AM, Cliff A, Romero J, Qin X, Mullins N, Docherty A, Coon H, Ruderfer DM; International Suicide Genetics Consortium; VA Million Veteran Program; MVP Suicide Exemplar Workgroup; Garvin MR, Pestian JP, Ashley-Koch AE, Beckham JC, McMahon B, Oslin DW, Kimbrel NA, Jacobson DA, Kainer D. Sullivan KA, et al. Commun Biol. 2024 Oct 21;7(1):1360. doi: 10.1038/s42003-024-06943-7. Commun Biol. 2024. PMID: 39433874 Free PMC article. - Genetics of disc-related disorders: current findings and lessons from other complex diseases.
Näkki A, Battié MC, Kaprio J. Näkki A, et al. Eur Spine J. 2014 Jun;23 Suppl 3:S354-63. doi: 10.1007/s00586-013-2878-2. Epub 2013 Jul 10. Eur Spine J. 2014. PMID: 23838702 Review. - Spag17 deficiency results in skeletal malformations and bone abnormalities.
Teves ME, Sundaresan G, Cohen DJ, Hyzy SL, Kajan I, Maczis M, Zhang Z, Costanzo RM, Zweit J, Schwartz Z, Boyan BD, Strauss JF 3rd. Teves ME, et al. PLoS One. 2015 May 27;10(5):e0125936. doi: 10.1371/journal.pone.0125936. eCollection 2015. PLoS One. 2015. PMID: 26017218 Free PMC article. - A COL11A2 mutation in Labrador retrievers with mild disproportionate dwarfism.
Frischknecht M, Niehof-Oellers H, Jagannathan V, Owczarek-Lipska M, Drögemüller C, Dietschi E, Dolf G, Tellhelm B, Lang J, Tiira K, Lohi H, Leeb T. Frischknecht M, et al. PLoS One. 2013;8(3):e60149. doi: 10.1371/journal.pone.0060149. Epub 2013 Mar 20. PLoS One. 2013. PMID: 23527306 Free PMC article. - Inferring positive selection in humans from genomic data.
Wollstein A, Stephan W. Wollstein A, et al. Investig Genet. 2015 Apr 1;6:5. doi: 10.1186/s13323-015-0023-1. eCollection 2015. Investig Genet. 2015. PMID: 25834723 Free PMC article.
References
- Galton F. Regression towards mediocrity in hereditary stature. J R Anthropol Inst. 1885;5:329–348.
- Fisher RA. The Correlation Between Relatives on the Supposition of Mendelian Inheritance. Transactions of the Royal Society of Edinburgh. 1918;52:399–433.
- Devlin B, Roeder K. Genomic control for association studies. Biometrics. 1999;55:997–1004. - PubMed
Publication types
MeSH terms
Grants and funding
- M01-RR00425/RR/NCRR NIH HHS/United States
- R01 MH061675/MH/NIMH NIH HHS/United States
- R01 CA050385/CA/NCI NIH HHS/United States
- 079895/WT_/Wellcome Trust/United Kingdom
- U01-HG004402/HG/NHGRI NIH HHS/United States
- R01 MH059587-09/MH/NIMH NIH HHS/United States
- N02-HL-6-4278/HL/NHLBI NIH HHS/United States
- R01 MH061675-09/MH/NIMH NIH HHS/United States
- R01 HL087679-01/HL/NHLBI NIH HHS/United States
- R01-MH67257/MH/NIMH NIH HHS/United States
- R01 DK073490/DK/NIDDK NIH HHS/United States
- K08 AR055688-03/AR/NIAMS NIH HHS/United States
- R01-HL088119/HL/NHLBI NIH HHS/United States
- R01-MH61675/MH/NIMH NIH HHS/United States
- 072856/WT_/Wellcome Trust/United Kingdom
- K23 DK080145/DK/NIDDK NIH HHS/United States
- N01-G65403/PHS HHS/United States
- R01 MH063706/MH/NIMH NIH HHS/United States
- CZB/4/279/CSO_/Chief Scientist Office/United Kingdom
- R01 AA007535/AA/NIAAA NIH HHS/United States
- R01 DK058845/DK/NIDDK NIH HHS/United States
- 081682/Z/06/Z/WT_/Wellcome Trust/United Kingdom
- R01 AA007535-08/AA/NIAAA NIH HHS/United States
- R01-MH60870/MH/NIMH NIH HHS/United States
- N01AG12100/AG/NIA NIH HHS/United States
- MH084698/MH/NIMH NIH HHS/United States
- R01 MH059565-06/MH/NIMH NIH HHS/United States
- R01 CA065725-14/CA/NCI NIH HHS/United States
- R56 DK062370/DK/NIDDK NIH HHS/United States
- N01-HC15103/HC/NHLBI NIH HHS/United States
- R01-HL59367/HL/NHLBI NIH HHS/United States
- R01 AA014041/AA/NIAAA NIH HHS/United States
- K05 AA017688/AA/NIAAA NIH HHS/United States
- N01HG65403/HG/NHGRI NIH HHS/United States
- R01 MH084698/MH/NIMH NIH HHS/United States
- N01HC55018/HL/NHLBI NIH HHS/United States
- N01-HC55021/HC/NHLBI NIH HHS/United States
- 079557/WT_/Wellcome Trust/United Kingdom
- R01 HL087647/HL/NHLBI NIH HHS/United States
- CZB/4/710/CSO_/Chief Scientist Office/United Kingdom
- U01 CA049449-21/CA/NCI NIH HHS/United States
- U01 HL084729/HL/NHLBI NIH HHS/United States
- U01 HL080295-04/HL/NHLBI NIH HHS/United States
- R01 AA014041-05/AA/NIAAA NIH HHS/United States
- N01-HC85086/HC/NHLBI NIH HHS/United States
- UL1 RR025005/RR/NCRR NIH HHS/United States
- R01-MH59571/MH/NIMH NIH HHS/United States
- AA13320/AA/NIAAA NIH HHS/United States
- R01-HL087676/HL/NHLBI NIH HHS/United States
- P01 CA087969/CA/NCI NIH HHS/United States
- U01 HL080295/HL/NHLBI NIH HHS/United States
- R01 HL087679/HL/NHLBI NIH HHS/United States
- N01-HC85079/HC/NHLBI NIH HHS/United States
- R01-MH59586/MH/NIMH NIH HHS/United States
- RL1-MH083268/MH/NIMH NIH HHS/United States
- R01 CA067262/CA/NCI NIH HHS/United States
- R01 HL087676/HL/NHLBI NIH HHS/United States
- 086596/WT_/Wellcome Trust/United Kingdom
- N01-HC85083/HC/NHLBI NIH HHS/United States
- R01 DA012854-09/DA/NIDA NIH HHS/United States
- R01 HG002651/HG/NHGRI NIH HHS/United States
- R01 DK075681-02/DK/NIDDK NIH HHS/United States
- HL69757/HL/NHLBI NIH HHS/United States
- U01-HL080295/HL/NHLBI NIH HHS/United States
- R01 HL043851/HL/NHLBI NIH HHS/United States
- R01 MH059571-05/MH/NIMH NIH HHS/United States
- N01-HC55222/HC/NHLBI NIH HHS/United States
- 088885/WT_/Wellcome Trust/United Kingdom
- R01 HL059367/HL/NHLBI NIH HHS/United States
- 075491/WT_/Wellcome Trust/United Kingdom
- Z01 AG000675/ImNIH/Intramural NIH HHS/United States
- 068545/Z/02/WT_/Wellcome Trust/United Kingdom
- R01-MH60879/MH/NIMH NIH HHS/United States
- R01 MH067257-04/MH/NIMH NIH HHS/United States
- DA12854/DA/NIDA NIH HHS/United States
- N01-HC55015/HC/NHLBI NIH HHS/United States
- R01-MH59588/MH/NIMH NIH HHS/United States
- R01-MH63706/MH/NIMH NIH HHS/United States
- R01-MH59565/MH/NIMH NIH HHS/United States
- N01-HC75150/HC/NHLBI NIH HHS/United States
- MC_QA137934/MRC_/Medical Research Council/United Kingdom
- K23 DK080145-01/DK/NIDDK NIH HHS/United States
- N01 HC015103/HC/NHLBI NIH HHS/United States
- 085301/WT_/Wellcome Trust/United Kingdom
- DK080145/DK/NIDDK NIH HHS/United States
- AA13321/AA/NIAAA NIH HHS/United States
- R01 MH060870-09/MH/NIMH NIH HHS/United States
- K99-HL094535/HL/NHLBI NIH HHS/United States
- U01 HG005214/HG/NHGRI NIH HHS/United States
- R01 MH060879-08/MH/NIMH NIH HHS/United States
- AA10248/AA/NIAAA NIH HHS/United States
- R01-HL086694/HL/NHLBI NIH HHS/United States
- R01 DK072193/DK/NIDDK NIH HHS/United States
- R01 HL086694/HL/NHLBI NIH HHS/United States
- DK58845/DK/NIDDK NIH HHS/United States
- U01 MH079469/MH/NIMH NIH HHS/United States
- U01 HL084756-03/HL/NHLBI NIH HHS/United States
- U01 MH079469-03/MH/NIMH NIH HHS/United States
- R01-HL087647/HL/NHLBI NIH HHS/United States
- R01 DK068336/DK/NIDDK NIH HHS/United States
- R01 DK091718/DK/NIDDK NIH HHS/United States
- PG/02/128/14470/BHF_/British Heart Foundation/United Kingdom
- MC_U127561128/MRC_/Medical Research Council/United Kingdom
- CA87969/CA/NCI NIH HHS/United States
- R01 AA013326/AA/NIAAA NIH HHS/United States
- T32 HG000040/HG/NHGRI NIH HHS/United States
- R01 HL059367-10/HL/NHLBI NIH HHS/United States
- U01-GM074518/GM/NIGMS NIH HHS/United States
- R56 DA012854/DA/NIDA NIH HHS/United States
- R01 HL087652/HL/NHLBI NIH HHS/United States
- G9521010(63660)/MRC_/Medical Research Council/United Kingdom
- G0600331(77796)/MRC_/Medical Research Council/United Kingdom
- R01 HL087652-01/HL/NHLBI NIH HHS/United States
- PG/02/128/BHF_/British Heart Foundation/United Kingdom
- RL1 MH083268-05/MH/NIMH NIH HHS/United States
- 085301/Z/08/Z/WT_/Wellcome Trust/United Kingdom
- F32 DK079466/DK/NIDDK NIH HHS/United States
- N01HC55022/HL/NHLBI NIH HHS/United States
- N01-HC55022/HC/NHLBI NIH HHS/United States
- CA49449/CA/NCI NIH HHS/United States
- U01 CA098233-08/CA/NCI NIH HHS/United States
- N01HC55222/HL/NHLBI NIH HHS/United States
- HHSN268200625226C/PHS HHS/United States
- U01 HG004402/HG/NHGRI NIH HHS/United States
- HG002651/HG/NHGRI NIH HHS/United States
- R01 DK075787/DK/NIDDK NIH HHS/United States
- R01 CA104021/CA/NCI NIH HHS/United States
- U01 CA067262/CA/NCI NIH HHS/United States
- R01 DK075681/DK/NIDDK NIH HHS/United States
- R01 CA065725/CA/NCI NIH HHS/United States
- R01-HL087641/HL/NHLBI NIH HHS/United States
- 077016/WT_/Wellcome Trust/United Kingdom
- R01-HL087652/HL/NHLBI NIH HHS/United States
- N01HC85086/HL/NHLBI NIH HHS/United States
- 083270/WT_/Wellcome Trust/United Kingdom
- R01 CA047988-20/CA/NCI NIH HHS/United States
- R01-DK075787/DK/NIDDK NIH HHS/United States
- R01 MH059588-08/MH/NIMH NIH HHS/United States
- N01HC55015/HL/NHLBI NIH HHS/United States
- R01-DK075681/DK/NIDDK NIH HHS/United States
- CA67262/CA/NCI NIH HHS/United States
- R01 HL087647-01/HL/NHLBI NIH HHS/United States
- R01 DK062370/DK/NIDDK NIH HHS/United States
- N01-HC55019/HC/NHLBI NIH HHS/United States
- R01 MH067257/MH/NIMH NIH HHS/United States
- 072960/WT_/Wellcome Trust/United Kingdom
- R01 AA013321/AA/NIAAA NIH HHS/United States
- K99 HL094535/HL/NHLBI NIH HHS/United States
- N01-HC85082/HC/NHLBI NIH HHS/United States
- DK072193/DK/NIDDK NIH HHS/United States
- R01 MH060870/MH/NIMH NIH HHS/United States
- R01 MH081800/MH/NIMH NIH HHS/United States
- F32 DK079466-01/DK/NIDDK NIH HHS/United States
- CA047988/CA/NCI NIH HHS/United States
- U01 HL084756/HL/NHLBI NIH HHS/United States
- HL084729/HL/NHLBI NIH HHS/United States
- P30 DK046200/DK/NIDDK NIH HHS/United States
- 076113/WT_/Wellcome Trust/United Kingdom
- U01-HG004399/HG/NHGRI NIH HHS/United States
- U01 DK062418/DK/NIDDK NIH HHS/United States
- P30 DK063491/DK/NIDDK NIH HHS/United States
- R01 MH059571/MH/NIMH NIH HHS/United States
- U01-MH79470/MH/NIMH NIH HHS/United States
- R01 MH059565/MH/NIMH NIH HHS/United States
- U01 GM074518-05/GM/NIGMS NIH HHS/United States
- RC2 HG005581-02/HG/NHGRI NIH HHS/United States
- R01 HL086694-02/HL/NHLBI NIH HHS/United States
- 079771/WT_/Wellcome Trust/United Kingdom
- G0600331/MRC_/Medical Research Council/United Kingdom
- R01 MH081800-01/MH/NIMH NIH HHS/United States
- HL71981/HL/NHLBI NIH HHS/United States
- 263-MA-410953/PHS HHS/United States
- P30 DK072488/DK/NIDDK NIH HHS/United States
- G9521010D/MRC_/Medical Research Council/United Kingdom
- Z01-HG000024/HG/NHGRI NIH HHS/United States
- R01 DK058845-11/DK/NIDDK NIH HHS/United States
- R01 DK068336-01/DK/NIDDK NIH HHS/United States
- R01-DK068336/DK/NIDDK NIH HHS/United States
- R01 CA050385-20/CA/NCI NIH HHS/United States
- R01-DK073490/DK/NIDDK NIH HHS/United States
- U01 MH079470-03/MH/NIMH NIH HHS/United States
- R01 DK075787-03/DK/NIDDK NIH HHS/United States
- U01 MH079470/MH/NIMH NIH HHS/United States
- R01 AG031890-02/AG/NIA NIH HHS/United States
- R01-MH59587/MH/NIMH NIH HHS/United States
- R01 DK072193-05/DK/NIDDK NIH HHS/United States
- N01-HC45133/HC/NHLBI NIH HHS/United States
- R01 MH059587/MH/NIMH NIH HHS/United States
- U01 HL072515-06/HL/NHLBI NIH HHS/United States
- U01-HL084756/HL/NHLBI NIH HHS/United States
- R01 HL088119-01/HL/NHLBI NIH HHS/United States
- Z01 HG000024/ImNIH/Intramural NIH HHS/United States
- 068545/WT_/Wellcome Trust/United Kingdom
- 090532/WT_/Wellcome Trust/United Kingdom
- U01-MH79469/MH/NIMH NIH HHS/United States
- 077016/Z/05/Z/WT_/Wellcome Trust/United Kingdom
- G0000649/MRC_/Medical Research Council/United Kingdom
- G9521010/MRC_/Medical Research Council/United Kingdom
- R01-AG031890/AG/NIA NIH HHS/United States
- N01HC55016/HL/NHLBI NIH HHS/United States
- U01 HL072515/HL/NHLBI NIH HHS/United States
- N01-HC55018/HC/NHLBI NIH HHS/United States
- G0701863/MRC_/Medical Research Council/United Kingdom
- DK079466/DK/NIDDK NIH HHS/United States
- UL1-RR025005/RR/NCRR NIH HHS/United States
- R01 DA012854/DA/NIDA NIH HHS/United States
- R01 HL071981-07/HL/NHLBI NIH HHS/United States
- N01-HC85081/HC/NHLBI NIH HHS/United States
- R01 MH059586/MH/NIMH NIH HHS/United States
- R01-MH59566/MH/NIMH NIH HHS/United States
- U01 CA098233/CA/NCI NIH HHS/United States
- 091746/WT_/Wellcome Trust/United Kingdom
- U01 DK062370-08/DK/NIDDK NIH HHS/United States
- AA014041/AA/NIAAA NIH HHS/United States
- R01 HL087700-03/HL/NHLBI NIH HHS/United States
- U54-RR020278/RR/NCRR NIH HHS/United States
- R01 MH059160-04/MH/NIMH NIH HHS/United States
- R01 CA047988/CA/NCI NIH HHS/United States
- R01 HL043851-10/HL/NHLBI NIH HHS/United States
- M01 RR000425/RR/NCRR NIH HHS/United States
- 064890/WT_/Wellcome Trust/United Kingdom
- U01 DK062370/DK/NIDDK NIH HHS/United States
- R01 MH059566-08/MH/NIMH NIH HHS/United States
- R01 HL071981/HL/NHLBI NIH HHS/United States
- R01-HL087700/HL/NHLBI NIH HHS/United States
- CZB/4/276/CSO_/Chief Scientist Office/United Kingdom
- G0500539/MRC_/Medical Research Council/United Kingdom
- N01AG12109/AG/NIA NIH HHS/United States
- G0000934/MRC_/Medical Research Council/United Kingdom
- G0601261/MRC_/Medical Research Council/United Kingdom
- N01-HC85085/HC/NHLBI NIH HHS/United States
- R01 DK089256/DK/NIDDK NIH HHS/United States
- U19 HL069757/HL/NHLBI NIH HHS/United States
- DK062370/DK/NIDDK NIH HHS/United States
- HG005214/HG/NHGRI NIH HHS/United States
- Z01 AG007380/ImNIH/Intramural NIH HHS/United States
- N01HC75150/HL/NHLBI NIH HHS/United States
- N01-HC85080/HC/NHLBI NIH HHS/United States
- K08-AR055688/AR/NIAMS NIH HHS/United States
- 086596/Z/08/Z/WT_/Wellcome Trust/United Kingdom
- N01HC25195/HL/NHLBI NIH HHS/United States
- 076113/C/04/Z/WT_/Wellcome Trust/United Kingdom
- R01 CA067262-14/CA/NCI NIH HHS/United States
- R01 HL087641-01/HL/NHLBI NIH HHS/United States
- T32-HG00040/HG/NHGRI NIH HHS/United States
- R01 AA013321-05/AA/NIAAA NIH HHS/United States
- DK063491/DK/NIDDK NIH HHS/United States
- R01 CA104021-02/CA/NCI NIH HHS/United States
- R01 MH059566/MH/NIMH NIH HHS/United States
- R01-MH059160/MH/NIMH NIH HHS/United States
- WT_/Wellcome Trust/United Kingdom
- R01 HG002651-05/HG/NHGRI NIH HHS/United States
- U01 HG005214-02/HG/NHGRI NIH HHS/United States
- CA50385/CA/NCI NIH HHS/United States
- HG005581/HG/NHGRI NIH HHS/United States
- K08 AR055688-04/AR/NIAMS NIH HHS/United States
- R01 DK073490-01/DK/NIDDK NIH HHS/United States
- P01 CA087969-12/CA/NCI NIH HHS/United States
- R01-MH79469/MH/NIMH NIH HHS/United States
- R01-HL087679/HL/NHLBI NIH HHS/United States
- N01-HC85084/HC/NHLBI NIH HHS/United States
- R01 CA049449/CA/NCI NIH HHS/United States
- RL1 MH083268/MH/NIMH NIH HHS/United States
- R01 MH059588/MH/NIMH NIH HHS/United States
- U01-CA098233/CA/NCI NIH HHS/United States
- N01-HC25195/HC/NHLBI NIH HHS/United States
- K08 AR055688/AR/NIAMS NIH HHS/United States
- R01-MH81800/MH/NIMH NIH HHS/United States
- U01 CA049449/CA/NCI NIH HHS/United States
- N01HC85079/HL/NHLBI NIH HHS/United States
- 084183/Z/07/Z/WT_/Wellcome Trust/United Kingdom
- R01 MH059586-08/MH/NIMH NIH HHS/United States
- R01 HL117078/HL/NHLBI NIH HHS/United States
- RC2 HG005581/HG/NHGRI NIH HHS/United States
- N01-HC55016/HC/NHLBI NIH HHS/United States
- R01 HL087641/HL/NHLBI NIH HHS/United States
- R01 AA013326-05/AA/NIAAA NIH HHS/United States
- U01 HL084729-03/HL/NHLBI NIH HHS/United States
- U01 HG004399-02/HG/NHGRI NIH HHS/United States
- N01 HC045133/HC/NHLBI NIH HHS/United States
- 091746/Z/10/Z/WT_/Wellcome Trust/United Kingdom
- R01 AA013320-04/AA/NIAAA NIH HHS/United States
- R01 HL087700/HL/NHLBI NIH HHS/United States
- MC_U106179471/MRC_/Medical Research Council/United Kingdom
- MC_U106188470/MRC_/Medical Research Council/United Kingdom
- U01 HL069757/HL/NHLBI NIH HHS/United States
- U01 HG004399/HG/NHGRI NIH HHS/United States
- U01 GM074518/GM/NIGMS NIH HHS/United States
- N01 HC035129/HC/NHLBI NIH HHS/United States
- U01-HL72515/HL/NHLBI NIH HHS/United States
- R01 AG031890/AG/NIA NIH HHS/United States
- U54 RR020278/RR/NCRR NIH HHS/United States
- 081682/WT_/Wellcome Trust/United Kingdom
- AA13326/AA/NIAAA NIH HHS/United States
- CRUK_/Cancer Research UK/United Kingdom
- K23-DK080145/DK/NIDDK NIH HHS/United States
- DK46200/DK/NIDDK NIH HHS/United States
- N01-HC35129/HC/NHLBI NIH HHS/United States
- 076113/B/04/Z/WT_/Wellcome Trust/United Kingdom
- R01 MH060879/MH/NIMH NIH HHS/United States
- U01 HL069757-10/HL/NHLBI NIH HHS/United States
- CA65725/CA/NCI NIH HHS/United States
- R01 AA013320/AA/NIAAA NIH HHS/United States
- N01-AG12100/AG/NIA NIH HHS/United States
- R01 HL087676-01/HL/NHLBI NIH HHS/United States
- U01 HG004402-02/HG/NHGRI NIH HHS/United States
- N01-HC55020/HC/NHLBI NIH HHS/United States
- R01 HL088119/HL/NHLBI NIH HHS/United States