Polyreactivity increases the apparent affinity of anti-HIV antibodies by heteroligation - PubMed (original) (raw)
. 2010 Sep 30;467(7315):591-5.
doi: 10.1038/nature09385.
Johannes F Scheid, Markus J Zoller, Michelle Krogsgaard, Rene G Ott, Shetha Shukair, Maxim N Artyomov, John Pietzsch, Mark Connors, Florencia Pereyra, Bruce D Walker, David D Ho, Patrick C Wilson, Michael S Seaman, Herman N Eisen, Arup K Chakraborty, Thomas J Hope, Jeffrey V Ravetch, Hedda Wardemann, Michel C Nussenzweig
Affiliations
- PMID: 20882016
- PMCID: PMC3699875
- DOI: 10.1038/nature09385
Polyreactivity increases the apparent affinity of anti-HIV antibodies by heteroligation
Hugo Mouquet et al. Nature. 2010.
Abstract
During immune responses, antibodies are selected for their ability to bind to foreign antigens with high affinity, in part by their ability to undergo homotypic bivalent binding. However, this type of binding is not always possible. For example, the small number of gp140 glycoprotein spikes displayed on the surface of the human immunodeficiency virus (HIV) disfavours homotypic bivalent antibody binding. Here we show that during the human antibody response to HIV, somatic mutations that increase antibody affinity also increase breadth and neutralizing potency. Surprisingly, the responding naive and memory B cells produce polyreactive antibodies, which are capable of bivalent heteroligation between one high-affinity anti-HIV-gp140 combining site and a second low-affinity site on another molecular structure on HIV. Although cross-reactivity to self-antigens or polyreactivity is strongly selected against during B-cell development, it is a common serologic feature of certain infections in humans, including HIV, Epstein-Barr virus and hepatitis C virus. Seventy-five per cent of the 134 monoclonal anti-HIV-gp140 antibodies cloned from six patients with high titres of neutralizing antibodies are polyreactive. Despite the low affinity of the polyreactive combining site, heteroligation demonstrably increases the apparent affinity of polyreactive antibodies to HIV.
Figures
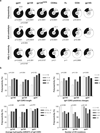
Figure 1. Polyreactivity, anti-cardiolipin and HEp-2 ELISAs
a, ELISAs measuring the reactivity of 4E10 (ref. 23), b12 (ref. 24), 2F5 (ref. 25) and 2G12 (ref. 26) against double-stranded DNA, single-stranded DNA, insulin and lipopolysaccharide. Dotted lines represent the positive control antibody ED38 (ref. 27). Horizontal lines show cut-off OD405 nm for positive reactivity. Green and red lines show the negative and low positive control antibodies, respectively,. b, As in a but for IgG antibodies cloned from gp140+ cells from patients 1–4 and for gp140− cells from patients 2 and 3. Pie charts summarize the frequency of polyreactive (black) and non-polyreactive (white) IgG+ clones, and for all pooled gp140+, gp140− and previously published control IgG antibodies (HC-IgGm) (right). P values are in comparison with pooled gp140− IgGs or between gp140+ and gp140− for patients 2 and 3 as indicated. c, Graphs show anti-cardiolipin ELISAs for the same antibodies as in b. Orange and blue lines show the positive control antibodies 4E10 and 2F5 (refs 23, 25), respectively. Pie charts summarize the frequency of cardiolipin reactive (black) and non-reactive (white) IgGs. P values are in comparison with pooled gp140− IgGs (right) or between gp140+ and gp140− for patients 2 and 3 as indicated. d, Same as b except for HEp-2 ELISAs.
Figure 2. IgH chain gene features and reactivity
a, Pie charts summarize polyreactivity, anti-cardiolipin and HEp-2 reactivity of gp140+ antibodies grouped by antibody specificity for gp41, gp120, gp120core, CD4bs, variable loops and CD4i. Reactive (black) and non-reactive (white) IgGs are shown; numbers in the centre indicate number of antibodies tested. P values are in comparison with gp140− IgGs unless otherwise indicated. b, Polyreactivity of all gp140-, gp120- and gp41-specific IgG antibodies grouped according to IgH CDR3 length, IgH CDR3-positive charges, IgH CDR3 hydropathy (GRAVY) or number of VH mutations.
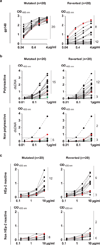
Figure 3. Reactivity of reverted antibodies
a, ELISAs against gp140 for 20 selected polyreactive and non-polyreactive antibodies (Supplementary Table 2), before (mutated) and after (reverted) reversion. The red line indicates the reactivity of the control b12 antibody. b, Anti-double-stranded DNA ELISAs (as example for polyreactivity) for the mutated and reverted antibodies that were initially polyreactive (top) or non-polyreactive (bottom). Numbers on the right indicate antibodies in reactive or non-reactive categories before and after reversion. c, As in b except for HEp-2 reactivity. Positive and negative control antibodies are as in Fig. 1.
Figure 4. Heteroligation
a, Representation of bivalent and monovalent homotypic binding versus heteroligation on high- and low-density gp120, high-density KLH and mixed chips. b, Apparent _K_a (_K_aapp) as measured by SPR on chips derivatized with high or low concentrations of gp120 or high concentrations of KLH, or a combination of low-gp120 and high-KLH. Numbers indicate the fold enhancement that can be attributed to heteroligation. See also Supplementary Table 4. Error bars, s.e.m. c, ELISA binding studies for IgG and corresponding Fabs on HIV-BaL. d, Solution competition-binding assay for fluorescent BaL virus using gp120 or KLH as competitors. Coloured lines show fitted curves. The y axis indicates the relative mean fluorescence index (rMFI), the x axis the competitor concentration in micromoles. Gp120 and KLH concentrations required to achieve 50% binding inhibition (IC50) are indicated. e, SPR sensorgrams for binding to BaL virus or gp120 protein to IgG antibody-immobilized sensor chips as illustrated by the schematic. RU, response units. f, Apparent _K_a (_K_aapp) for gp120 protein as measured by SPR. Error bars, s.e.m. g, As f but for the relative _K_a to BaL virus (_K_arelBaL).
Comment in
- HIV: Antibodies with a split personality.
Plückthun A. Plückthun A. Nature. 2010 Sep 30;467(7315):537-8. doi: 10.1038/467537a. Nature. 2010. PMID: 20882002 No abstract available.
Similar articles
- HIV: Antibodies with a split personality.
Plückthun A. Plückthun A. Nature. 2010 Sep 30;467(7315):537-8. doi: 10.1038/467537a. Nature. 2010. PMID: 20882002 No abstract available. - Memory B cell antibodies to HIV-1 gp140 cloned from individuals infected with clade A and B viruses.
Mouquet H, Klein F, Scheid JF, Warncke M, Pietzsch J, Oliveira TY, Velinzon K, Seaman MS, Nussenzweig MC. Mouquet H, et al. PLoS One. 2011;6(9):e24078. doi: 10.1371/journal.pone.0024078. Epub 2011 Sep 8. PLoS One. 2011. PMID: 21931643 Free PMC article. - Identification and characterization of a new cross-reactive human immunodeficiency virus type 1-neutralizing human monoclonal antibody.
Zhang MY, Xiao X, Sidorov IA, Choudhry V, Cham F, Zhang PF, Bouma P, Zwick M, Choudhary A, Montefiori DC, Broder CC, Burton DR, Quinnan GV Jr, Dimitrov DS. Zhang MY, et al. J Virol. 2004 Sep;78(17):9233-42. doi: 10.1128/JVI.78.17.9233-9242.2004. J Virol. 2004. PMID: 15308718 Free PMC article. - HIV Broadly Neutralizing Antibodies: VRC01 and Beyond.
Wu X. Wu X. Adv Exp Med Biol. 2018;1075:53-72. doi: 10.1007/978-981-13-0484-2_3. Adv Exp Med Biol. 2018. PMID: 30030789 Review. - Polyreactive Antibodies in Anti-HIV-1 Responses.
Liao H, Zhang Z. Liao H, et al. Curr Mol Med. 2018;18(2):126-133. doi: 10.2174/1566524018666180720165406. Curr Mol Med. 2018. PMID: 30033868 Review.
Cited by
- Responsiveness of B cells is regulated by the hinge region of IgD.
Übelhart R, Hug E, Bach MP, Wossning T, Dühren-von Minden M, Horn AH, Tsiantoulas D, Kometani K, Kurosaki T, Binder CJ, Sticht H, Nitschke L, Reth M, Jumaa H. Übelhart R, et al. Nat Immunol. 2015 May;16(5):534-43. doi: 10.1038/ni.3141. Epub 2015 Apr 6. Nat Immunol. 2015. PMID: 25848865 - Mixed adjuvant formulations reveal a new combination that elicit antibody response comparable to Freund's adjuvants.
Lai RP, Seaman MS, Tonks P, Wegmann F, Seilly DJ, Frost SD, LaBranche CC, Montefiori DC, Dey AK, Srivastava IK, Sattentau Q, Barnett SW, Heeney JL. Lai RP, et al. PLoS One. 2012;7(4):e35083. doi: 10.1371/journal.pone.0035083. Epub 2012 Apr 11. PLoS One. 2012. PMID: 22509385 Free PMC article. - Heads, stalks and everything else: how can antibodies eradicate influenza as a human disease?
Neu KE, Henry Dunand CJ, Wilson PC. Neu KE, et al. Curr Opin Immunol. 2016 Oct;42:48-55. doi: 10.1016/j.coi.2016.05.012. Epub 2016 Jun 3. Curr Opin Immunol. 2016. PMID: 27268395 Free PMC article. Review. - Transmembrane domain membrane proximal external region but not surface unit-directed broadly neutralizing HIV-1 antibodies can restrict dendritic cell-mediated HIV-1 trans-infection.
Sagar M, Akiyama H, Etemad B, Ramirez N, Freitas I, Gummuluru S. Sagar M, et al. J Infect Dis. 2012 Apr 15;205(8):1248-57. doi: 10.1093/infdis/jis183. Epub 2012 Mar 6. J Infect Dis. 2012. PMID: 22396600 Free PMC article. - Improved killing of HIV-infected cells using three neutralizing and non-neutralizing antibodies.
Tuyishime M, Garrido C, Jha S, Moeser M, Mielke D, LaBranche C, Montefiori D, Haynes BF, Joseph S, Margolis DM, Ferrari G. Tuyishime M, et al. J Clin Invest. 2020 Oct 1;130(10):5157-5170. doi: 10.1172/JCI135557. J Clin Invest. 2020. PMID: 32584790 Free PMC article.
References
- Zhu P, et al. Distribution and three-dimensional structure of AIDS virus envelope spikes. Nature. 2006;441:847–852. - PubMed
- Wardemann H, et al. Predominant autoantibody production by early human B cell precursors. Science. 2003;301:1374–1377. - PubMed
- Scheid JF, et al. Broad diversity of neutralizing antibodies isolated from memory B cells in HIV-infected individuals. Nature. 2009;458:636–640. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P01 AI081677/AI/NIAID NIH HHS/United States
- R01 AI047770/AI/NIAID NIH HHS/United States
- 1 P01 AI081677/AI/NIAID NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources