An insect herbivore microbiome with high plant biomass-degrading capacity - PubMed (original) (raw)
. 2010 Sep 23;6(9):e1001129.
doi: 10.1371/journal.pgen.1001129.
Jarrod J Scott, Frank O Aylward, Sandra M Adams, Susannah G Tringe, Adrián A Pinto-Tomás, Clifton E Foster, Markus Pauly, Paul J Weimer, Kerrie W Barry, Lynne A Goodwin, Pascal Bouffard, Lewyn Li, Jolene Osterberger, Timothy T Harkins, Steven C Slater, Timothy J Donohue, Cameron R Currie
Affiliations
- PMID: 20885794
- PMCID: PMC2944797
- DOI: 10.1371/journal.pgen.1001129
An insect herbivore microbiome with high plant biomass-degrading capacity
Garret Suen et al. PLoS Genet. 2010.
Abstract
Herbivores can gain indirect access to recalcitrant carbon present in plant cell walls through symbiotic associations with lignocellulolytic microbes. A paradigmatic example is the leaf-cutter ant (Tribe: Attini), which uses fresh leaves to cultivate a fungus for food in specialized gardens. Using a combination of sugar composition analyses, metagenomics, and whole-genome sequencing, we reveal that the fungus garden microbiome of leaf-cutter ants is composed of a diverse community of bacteria with high plant biomass-degrading capacity. Comparison of this microbiome's predicted carbohydrate-degrading enzyme profile with other metagenomes shows closest similarity to the bovine rumen, indicating evolutionary convergence of plant biomass degrading potential between two important herbivorous animals. Genomic and physiological characterization of two dominant bacteria in the fungus garden microbiome provides evidence of their capacity to degrade cellulose. Given the recent interest in cellulosic biofuels, understanding how large-scale and rapid plant biomass degradation occurs in a highly evolved insect herbivore is of particular relevance for bioenergy.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
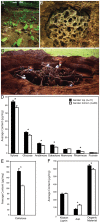
Figure 1. Organic polymer characterization of leaf-cutter ant fungus gardens.
Leaf-cutter ants forage for leaves (A) that they use to cultivate a fungus in specialized gardens (B) within their massive colonies (C). Sugar composition analysis of the plant biomass from the top and bottom layers of multiple fungus garden chambers shows an overall decrease in average content for many of the components of hemicellulose (D) and cellulose (E). In contrast, lignin (F) exhibited no change in average content. Error bars in graphs are standard error of the mean. The asterisks indicate a significant decrease in overall average content between top and bottom samples (two-tailed paired t test, P<0.05). [Photo credits: river of leaves, used under the GNU Free Documentation License CC-BY-SA-3.0,2.5,2.0,1.0; exposed fungus garden, Jarrod J. Scott/University of Wisconsin-Madison; concrete nest, Wolfang Thaler].
Figure 2. Phylogenetic analysis of the leaf-cutter ant fungus garden.
A phylogenetic analysis of near-full length 16S rDNA sequence libraries from the top (A) and bottom (B) layers of leaf-cutter ant fungus gardens was performed. Identified phylotypes were tabulated and mapped to their respective phyla as shown. Total numbers of phylotypes are shown to the right of each phylum, and the total number of clones for each phylum is shown in square brackets. Comparison of top and bottom layers indicates that leaf-cutter ant fungus gardens are dominated by phylotypes belonging to the α-proteobacteria, β-proteobacteria, γ-proteobacteria, Actinobacteria, and the Bacteroidetes as highlighted. Phylotypes belonging to specific phyla were found exclusive to top and bottom samples, including the Gemmatimonadetes and candidate phylum SPAM (blue lettering) in the top, and the Chloroflexi and candidate phylum TM7 (red lettering) in the bottom of the garden.
Figure 3. CAZy clustering of the fungus garden metagenome.
Comparative clustering of the leaf-cutter ant fungus garden community metagenome with 13 other metagenomes. The predicted proteome from each metagenome was compared using carbohydrate-active enzymes (CAZy) profiles (A) and clusters of orthologous groups (COGs) profiles (B). CAZy and COG profiles for each metagenome was generated and clustered using Pearson's product moment. An unrooted tree (UPGMA) was then generated using PHYLIP and visualized using phylodendron.
Figure 4. Leaf-cutter ant fungus garden metagenome recruitment analysis.
Leaf-cutter ant fungus garden metagenome recruitment analysis. Reads from the leaf-cutter ant fungus garden community metagenome are shown mapped onto the draft genome sequences of the two leaf-cutter ant bacterial symbionts Klebsiella variicola At-22 (A) and Pantoea sp. At-9b (B). The sequence identity of each recruited read is as follows: blue, 95%–100%; magenta, 90%–95%; yellow, 85%–90%; gold, 80%–85%, and red, 75%–80%. The draft genomes are represented as concatenated contigs in order of decreasing size, and the corresponding coordinates are shown in the second-most inner ring. The average GC content for these draft genomes are shown in the innermost ring with green representing above-average GC content, and olive representing below-average GC content.
References
- Sticklen MB. Plant genetic engineering for biofuel production: towards affordable cellulosic ethanol. Nat Rev Genet. 2008;9:433–443. - PubMed
- Warnecke F, Luginbuhl P, Ivanova N, Ghassemian M, Richardson TH, et al. Metagenomic and functional analysis of hindgut microbiota of a wood-feeding higher termite. Nature. 2007;450:560–565. - PubMed
- Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, et al. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–1031. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases