Recombinant E. coli prototype strains for in vivo glycorandomization - PubMed (original) (raw)
Recombinant E. coli prototype strains for in vivo glycorandomization
Gavin J Williams et al. ACS Chem Biol. 2011.
Abstract
In vitro glycorandomization is a powerful strategy to alter the glycosylation patterns of natural products and small molecule therapeutics. Yet, such in vitro methods are often difficult to scale and can be costly given the requirement to provide various nucleotides and cofactors. Here, we report the construction of several recombinant E. coli prototype strains that allow the facile production of a range of small molecule glycosides. This strategy relies on the engineered promiscuity of three key enzymes, an anomeric kinase, a sugar-1-phosphate nucleotidyltransferase, and a glycosyltransferase, as well as the ability of diverse small molecules to freely enter E. coli. Subsequently, this work is the first demonstration of "in vivo glycorandomization" and offers vast combinatorial potential by simple fermentation.
Figures
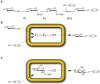
Figure 1
Comparison of methods for glycodiversification of natural products. (a) In vitro glycorandomization. Reducing sugars are converted to sugar-1-phosphates by E1, a flexible anomeric kinase. E2, A suitably flexible sugar-1-phosphate nucleotidyltransferase activates each sugar phosphate to the corresponding nucleotide sugar. Large panels of NDP-donors are used to probe the specificity of natural product GTs. Grey oval represents diverse natural product or natural product-like aglycons (X = O, S, or NH). (b) In vivo glycodiversification via a ‘non-natural glycoside host’ strain. Reducing sugars and aglycons are fed to a bacterial host engineered to express E1, E2, and a promiscuous GT. The endogenous biosynthetic machinery ensures recycling of necessary cofactors and aglycons decorated with non-natural sugars are collected from the culture media. (c) In vivo glucoside host. Aglycons are fed into a bacterial host engineered to express a GT which uses endogenous dTDP/UDPGlc as the glycosyl donor.
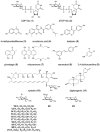
Figure 2
Structures of substrates used in this study.
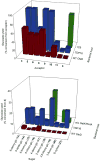
Figure 3
Activity of prototype glycoside producing strains. (a) Yields (% conversion from acceptor) of glucosides using the TDP16-, WT-, and 1C9-based glucoside host with a small panel of diverse acceptors. (b) Yields (% conversion from 3) of glycosides using the TDP16- and WT-based non-natural glycoside host using acceptor 3 and a panel of free sugars. ‘w/o GalK/RmlA’ refers to the TDP16-based host but which lacks the pDuet-GalK/RmlA vector. See Supplementary Materials for full description of the strains used, details of bioconversion conditions and detection. The standard deviation of the % conversions using data from three independent determinations was less than 20%.
Comment in
- Fermenting next generation glycosylated therapeutics.
Chen X. Chen X. ACS Chem Biol. 2011 Jan 21;6(1):14-7. doi: 10.1021/cb100375y. ACS Chem Biol. 2011. PMID: 21250649
Similar articles
- Fermenting next generation glycosylated therapeutics.
Chen X. Chen X. ACS Chem Biol. 2011 Jan 21;6(1):14-7. doi: 10.1021/cb100375y. ACS Chem Biol. 2011. PMID: 21250649 - Expanding the promiscuity of a natural-product glycosyltransferase by directed evolution.
Williams GJ, Zhang C, Thorson JS. Williams GJ, et al. Nat Chem Biol. 2007 Oct;3(10):657-62. doi: 10.1038/nchembio.2007.28. Epub 2007 Sep 9. Nat Chem Biol. 2007. PMID: 17828251 - Structure-based engineering of E. coli galactokinase as a first step toward in vivo glycorandomization.
Yang J, Fu X, Liao J, Liu L, Thorson JS. Yang J, et al. Chem Biol. 2005 Jun;12(6):657-64. doi: 10.1016/j.chembiol.2005.04.009. Chem Biol. 2005. PMID: 15975511 - Neoglycorandomization and chemoenzymatic glycorandomization: two complementary tools for natural product diversification.
Langenhan JM, Griffith BR, Thorson JS. Langenhan JM, et al. J Nat Prod. 2005 Nov;68(11):1696-711. doi: 10.1021/np0502084. J Nat Prod. 2005. PMID: 16309329 Review. - Biotechnological advances in UDP-sugar based glycosylation of small molecules.
De Bruyn F, Maertens J, Beauprez J, Soetaert W, De Mey M. De Bruyn F, et al. Biotechnol Adv. 2015 Mar-Apr;33(2):288-302. doi: 10.1016/j.biotechadv.2015.02.005. Epub 2015 Feb 16. Biotechnol Adv. 2015. PMID: 25698505 Review.
Cited by
- Advances in glycosyltransferase-mediated glycodiversification of small molecules.
Thuan NH, Huong QTT, Lam BD, Tam HT, Thu PT, Canh NX, Tatipamula VB. Thuan NH, et al. 3 Biotech. 2024 Sep;14(9):209. doi: 10.1007/s13205-024-04044-0. Epub 2024 Aug 23. 3 Biotech. 2024. PMID: 39184913 Review. - Expanding the Substrate Scope of a Bacterial Nucleotidyltransferase via Allosteric Mutations.
Zheng M, Zheng M, Lupoli TJ. Zheng M, et al. ACS Infect Dis. 2022 Oct 14;8(10):2035-2044. doi: 10.1021/acsinfecdis.2c00402. Epub 2022 Sep 15. ACS Infect Dis. 2022. PMID: 36106727 Free PMC article. - Metabolic labeling of the bacterial peptidoglycan by functionalized glucosamine.
Xu Y, Hernández-Rocamora VM, Lorent JH, Cox R, Wang X, Bao X, Stel M, Vos G, van den Bos RM, Pieters RJ, Gray J, Vollmer W, Breukink E. Xu Y, et al. iScience. 2022 Jul 12;25(8):104753. doi: 10.1016/j.isci.2022.104753. eCollection 2022 Aug 19. iScience. 2022. PMID: 35942089 Free PMC article. - Exploring the broad nucleotide triphosphate and sugar-1-phosphate specificity of thymidylyltransferase Cps23FL from Streptococcus pneumonia serotype 23F.
Li S, Wang H, Jin G, Chen Z, Gu G. Li S, et al. RSC Adv. 2020 Aug 14;10(50):30110-30114. doi: 10.1039/d0ra05799a. eCollection 2020 Aug 10. RSC Adv. 2020. PMID: 35518267 Free PMC article. - Sugar-Pirating as an Enabling Platform for the Synthesis of 4,6-Dideoxyhexoses.
Zhang Y, Zhang J, Ponomareva LV, Cui Z, Van Lanen SG, Thorson JS. Zhang Y, et al. J Am Chem Soc. 2020 May 20;142(20):9389-9395. doi: 10.1021/jacs.9b13766. Epub 2020 May 7. J Am Chem Soc. 2020. PMID: 32330028 Free PMC article.
References
- Williams GJ, Zhang C, Thorson JS. Natural product glycosyltransferases: Properties and applications. Adv Enzymol Relat Areas Mol Biol. 2008;76:55–119. - PubMed
- Williams GJ, Zhang C, Thorson JS. Expanding the promiscuity of a natural-product glycosyltransferase by directed evolution. Nat Chem Biol. 2007;3:657–62. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R37 AI052218/AI/NIAID NIH HHS/United States
- R01 AI052218-10/AI/NIAID NIH HHS/United States
- R01 AI052218/AI/NIAID NIH HHS/United States
- AI52218/AI/NIAID NIH HHS/United States
- R01 AI052218-08/AI/NIAID NIH HHS/United States
- R01 AI052218-09/AI/NIAID NIH HHS/United States
- R01 AI052218-07/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources