IFI16 is an innate immune sensor for intracellular DNA - PubMed (original) (raw)
. 2010 Nov;11(11):997-1004.
doi: 10.1038/ni.1932. Epub 2010 Oct 3.
Sinead E Keating, Marcin Baran, Kristy A Horan, Søren B Jensen, Shruti Sharma, Cherilyn M Sirois, Tengchuan Jin, Eicke Latz, T Sam Xiao, Katherine A Fitzgerald, Søren R Paludan, Andrew G Bowie
Affiliations
- PMID: 20890285
- PMCID: PMC3142795
- DOI: 10.1038/ni.1932
IFI16 is an innate immune sensor for intracellular DNA
Leonie Unterholzner et al. Nat Immunol. 2010 Nov.
Abstract
The detection of intracellular microbial DNA is critical to appropriate innate immune responses; however, knowledge of how such DNA is sensed is limited. Here we identify IFI16, a PYHIN protein, as an intracellular DNA sensor that mediates the induction of interferon-β (IFN-β). IFI16 directly associated with IFN-β-inducing viral DNA motifs. STING, a critical mediator of IFN-β responses to DNA, was recruited to IFI16 after DNA stimulation. Lowering the expression of IFI16 or its mouse ortholog p204 by RNA-mediated interference inhibited gene induction and activation of the transcription factors IRF3 and NF-κB induced by DNA and herpes simplex virus type 1 (HSV-1). IFI16 (p204) is the first PYHIN protein to our knowledge shown to be involved in IFN-β induction. Thus, the PYHIN proteins IFI16 and AIM2 form a new family of innate DNA sensors we call 'AIM2-like receptors' (ALRs).
Figures
Figure 1
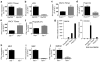
Induction of IFNβ by a VACV DNA motif. a, HEK293T cells were transfected with nucleic acids (0.5 and 5 μg/ml) for 16 h and IFNβ promoter activity was measured by reporter gene assay. b, PMA-treated THP-1 cells were transfected with 1 μg/ml poly(dA-dT) or with 200 ng/ml DNA isolated from VACV, calf thymus or Listeria for 6 h and IFNβ mRNA was measured. c, d THP1 (c) or immortalized MEF (d) cells were transfected with 1 μg/ml (c) or 5 μg/ml (d) nucleic acids for 6 h and IFNβ mRNA was measured. e, HEK293T cells were transfected with 5 μg/ml nucleic acids for 16 h, and IFNβ promoter activity was measured. f, g, THP-1 (f) or RAW264.7 (g) cells were transfected with different lengths (b.p.) of 1 μg/ml VACV 70mer-derived DNA, GC-rich 70mer (70(GC)) or interferon stimulatory DNA (ISD) for 6 h and IFNβ mRNA was measured. Error bars indicate s.d.
Figure 2
Induction of IFNβ by a VACV DNA motif is independent of known DNA sensing pathways. a-f, h-j, Immortalised BMDMs or MEFs derived from mice lacking signalling components as indicated were transfected with 5 μg/ml VACV 70mer (a, c, e, h-j) or poly(dA-dT) (f) or stimulated with LPS (b) or poly(I:C) (d) for 6 h and IFNβ mRNA measured. Values are expressed as % of stimulation observed in cells from wild type mice. Data is from three independent experiments performed in triplicate and error bars represent s.e.m. g, Immortalised BMDMs were transfected with nucleic acid for 6h and IFNβ mRNA measured (left panel). 100 ng of the RNA extracted from BMDMs was transfected into HEK293T cells for 16 h, and IFNβ promoter activity measured (right panel). Data from one experiment of three is shown where error bars represent s.d.
Figure 3
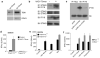
IFI16 binds to immune stimulatory viral DNA. a, Schematic representation of human PYHINs. Aa, amino acids. b, Cytoplasmic extracts from PMA-treated THP-1 cells were incubated with biotinylated ss or dsVACV 70mer immobilised on streptavidin beads. Precipitated proteins were immunoblotted with an IFI16-specific antibody as indicated. c, Left panel: THP-1 cells were transfected with 1 μg/ml dsVACV 70mer or poly(I:C) in the absence or presence of 1 μg/ml ssVACV 70mer and IFNβ mRNA was measured after 6 h. Right panel: HEK293T cells were transfected with 50 ng/ml poly(dA-dT) alone, or together with 50 ng/ml ss or dsVACV 70mer and IFNβ promoter activity measured after 16 h. Data from one experiment of three is shown where error bars represent s.d. d, PMA/IFN α-treated THP-1 cells grown on coverslips were transfected with 2.5 μg/ml VACV 70mer or poly(I:C) for 1 h. Cells were fixed and stained with anti-IFI16 antibody (red). DNA and poly(I:C) were visualised with DAPI (blue). e, FITC-labelled HSV-1 60mer was transfected into PMA-treated THP-1 cells for 3 h. Cells were fixed and stained for IFI16 (red). DAPI-stained DNA is shown in blue (upper panel) and the HSV 60mer is shown in green (lower panel). Scale bar: 10 μm. f, AlphaScreen assessment of IFI16 HIN domains binding dsVACV 70mer. Left panel, 30 nM DNA with increasing concentration of HIN domains or GB1 expression tag. Right panel, 30 nM protein domain with increasing concentrations of biotin-70mer.
Figure 4
Role for STING in IFI16-mediated IFNβ induction. a, Cytoplasmic extracts from PMA-treated THP-1 cells were incubated with biotinylated ss or dsVACV 70mer immobilised on streptavidin beads. Precipitated proteins were immunoblotted as indicated. b, PMA/IFNα pre-treated THP1 cells were transfected with 1 μg/ml VACV 70mer for 4 h. Resulting lysates were incubated with anti-STING antibody and immuno-precipitated proteins were immunoblotted as indicated. c, Myc-tagged human (hu) or mouse (mur) STING were overexpressed in HEK293 cells, immobilized on sepharose beads, and incubated with lysates from DNA-treated THP1 cells. Co-immunoprecipitated IFI16 was detected by immunoblotting. Ab, antibody heavy chain. d, e, BMDMs lacking STING were transfected with either VACV 70mer , HSV 60mer or infected with HSV-1 or Sendai virus for 18 h as indicated and IFNβ protein release was measured by ELISA. f, Immortalised BMDMs from ASC-null mice were transfected with DNA as indicated for 18 h and IFNβ protein secretion was measured by ELISA. Data from one experiment of at least 2 (performed in triplicate) is shown where error bars represent s.d. (d-f).
Figure 5
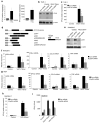
IFI16/p204 is required for DNA-mediated gene induction. a, Left panel, IFI16 mRNA was measured in THP-1, PMA-treated THP-1, or HEK293 cells. Right panel, THP-1 or PMA-treated THP-1 cells were transfected with VACV 70mer for 6 h and IFNβ mRNA measured. b, c, siRNA-treated THP-1 cells were transfected with 1 μg/ml VACV 70mer for 6 h, before detection of IFI16 protein expression by immunoblotting (b) or measurement of IFNβ mRNA (c). d, Domain organization of human IFI16 compared to murine members of the PYHIN family, drawn to scale. Boxes represent conserved domains. aa, amino acids. Of the murine proteins, p204 (*) is most similar to human IFI16, in terms of domain structure and aa identity. e, f, siRNA-treated RAW264.7 cells were transfected with 1 μg/ml VACV 70mer for 6 h and p204 protein expression was analysed by immunoblotting (e) or p204, IFNβ, CCL5 and TNF mRNA was measured (f). g, siRNA-treated MEF cells were transfected with 1 μg/ml VACV 70mer for 6 h and p204, IFNβ, CCL5 and TNF mRNA was measured. h, siRNA-treated RAW264.7 cells were treated with HSV 60mer DNA for 6 h before measurement of IFNβ mRNA. i, BMDMs were electroporated with siRNA prior to transfection with oligomers for 18 h, and IFNβ protein release was measured. Data from one experiment of 2-3 (performed in triplicate) is shown (a, c, f-i) where error bars represent s.d. *P < 0.05 compared with control siRNA.
Figure 6
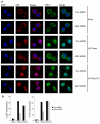
p204 is required for VACV 70mer DNA-stimulated transcription factor activation. a, siRNA-treated RAW264.7 cells grown on glass coverslips were mock-transfected or transfected with 2.5 μg/ml VACV 70mer or poly(I:C) for 6 h, fixed and stained for NF-κB p65 (red) and IRF3 (green). Nuclei were visualised by DAPI (blue). b, Cells treated as in (a) were qualitatively examined to assess staining of either p65 (left panel) or IRF3 (right panel) in the nucleus. Cells showing nuclear staining were counted and expressed as a percentage of total number of cells. At least 200 cells were counted per sample. Shown is a representative of four independent experiments.
Figure 7
IFI16/p204 is required for the innate immune response to HSV-1. a, BMDMs were infected with HSV-1 (MOI=10) for 6 h and IFNβ mRNA was measured. b, RAW264.7 cells were pre-treated with ML60812 for 2 h prior to infection with HSV-1 (MOI=10), or transfection with poly(dA-dT) for 6 h. c, d, siRNA-treated RAW264.7 cells were infected with HSV-1 or Sendai virus for 6 h and IFNβ (c, d), CXCL10 (c), IL-6 (c) and TNF (c) mRNA was measured. e, siRNA-treated RAW264.7 cells were infected with HSV1 (MOI 10) or Sendai virus for 20 h and IFNβ protein expression determined by ELISA. f, siRNA-treated RAW264.7 cells grown on glass coverslips were mock-infected or infected with HSV-1 or Sendai virus for 6 h, fixed and stained for NF-κB p65 (red) and IRF3 (green). Nuclei were visualised by DAPI (blue). g, Cells treated as in (f) were qualitatively examined to assess staining of either p65 (left panel) or IRF3 (right panel) in the nucleus. Cells showing nuclear staining were counted and expressed as a percentage of total number of cells. At least 200 cells were counted per sample. Shown is a representative of three independent experiments. h, RAW264.7 cells transfected with HSV 60mer (2 μg/ml) were infected with HSV-1 (MOI 1) and culture supernatants were harvested at the indicated time points post infection for viral quantification by plaque assay. Data from one experiment of 2-3 (performed in triplicate) is shown (a-e, h) where error bars represent s.d.*P < 0.001 compared with control siRNA.
Comment in
- PYHIN proteins: center stage in DNA sensing.
Goubau D, Rehwinkel J, Reis e Sousa C. Goubau D, et al. Nat Immunol. 2010 Nov;11(11):984-6. doi: 10.1038/ni1110-984. Nat Immunol. 2010. PMID: 20959802 No abstract available.
Similar articles
- BRCA1 Regulates IFI16 Mediated Nuclear Innate Sensing of Herpes Viral DNA and Subsequent Induction of the Innate Inflammasome and Interferon-β Responses.
Dutta D, Dutta S, Veettil MV, Roy A, Ansari MA, Iqbal J, Chikoti L, Kumar B, Johnson KE, Chandran B. Dutta D, et al. PLoS Pathog. 2015 Jun 29;11(6):e1005030. doi: 10.1371/journal.ppat.1005030. eCollection 2015 Jun. PLoS Pathog. 2015. PMID: 26121674 Free PMC article. Retracted. - The intracellular DNA sensors cGAS and IFI16 do not mediate effective antiviral immune responses to HSV-1 in human microglial cells.
Jeffries AM, Nitika, Truman AW, Marriott I. Jeffries AM, et al. J Neurovirol. 2020 Aug;26(4):544-555. doi: 10.1007/s13365-020-00852-1. Epub 2020 Jun 2. J Neurovirol. 2020. PMID: 32488842 Free PMC article. - Histone H2B-IFI16 Recognition of Nuclear Herpesviral Genome Induces Cytoplasmic Interferon-β Responses.
Iqbal J, Ansari MA, Kumar B, Dutta D, Roy A, Chikoti L, Pisano G, Dutta S, Vahedi S, Veettil MV, Chandran B. Iqbal J, et al. PLoS Pathog. 2016 Oct 20;12(10):e1005967. doi: 10.1371/journal.ppat.1005967. eCollection 2016 Oct. PLoS Pathog. 2016. PMID: 27764250 Free PMC article. - The interferon-inducible DNA-sensor protein IFI16: a key player in the antiviral response.
Dell'Oste V, Gatti D, Giorgio AG, Gariglio M, Landolfo S, De Andrea M. Dell'Oste V, et al. New Microbiol. 2015 Jan;38(1):5-20. Epub 2015 Jan 1. New Microbiol. 2015. PMID: 25742143 Review. - Interferon-inducible p200-family protein IFI16, an innate immune sensor for cytosolic and nuclear double-stranded DNA: regulation of subcellular localization.
Veeranki S, Choubey D. Veeranki S, et al. Mol Immunol. 2012 Jan;49(4):567-71. doi: 10.1016/j.molimm.2011.11.004. Epub 2011 Dec 2. Mol Immunol. 2012. PMID: 22137500 Free PMC article. Review.
Cited by
- Constitutive interferon-inducible protein 16-inflammasome activation during Epstein-Barr virus latency I, II, and III in B and epithelial cells.
Ansari MA, Singh VV, Dutta S, Veettil MV, Dutta D, Chikoti L, Lu J, Everly D, Chandran B. Ansari MA, et al. J Virol. 2013 Aug;87(15):8606-23. doi: 10.1128/JVI.00805-13. Epub 2013 May 29. J Virol. 2013. PMID: 23720728 Free PMC article. - The Trinity of cGAS, TLR9, and ALRs Guardians of the Cellular Galaxy Against Host-Derived Self-DNA.
Kumar V. Kumar V. Front Immunol. 2021 Feb 11;11:624597. doi: 10.3389/fimmu.2020.624597. eCollection 2020. Front Immunol. 2021. PMID: 33643304 Free PMC article. Review. - A proteomics perspective on viral DNA sensors in host defense and viral immune evasion mechanisms.
Crow MS, Javitt A, Cristea IM. Crow MS, et al. J Mol Biol. 2015 Jun 5;427(11):1995-2012. doi: 10.1016/j.jmb.2015.02.016. Epub 2015 Feb 26. J Mol Biol. 2015. PMID: 25728651 Free PMC article. Review. - Mst1 shuts off cytosolic antiviral defense through IRF3 phosphorylation.
Meng F, Zhou R, Wu S, Zhang Q, Jin Q, Zhou Y, Plouffe SW, Liu S, Song H, Xia Z, Zhao B, Ye S, Feng XH, Guan KL, Zou J, Xu P. Meng F, et al. Genes Dev. 2016 May 1;30(9):1086-100. doi: 10.1101/gad.277533.116. Epub 2016 Apr 28. Genes Dev. 2016. PMID: 27125670 Free PMC article. - DNA recognition in immunity and disease.
Holm CK, Paludan SR, Fitzgerald KA. Holm CK, et al. Curr Opin Immunol. 2013 Feb;25(1):13-8. doi: 10.1016/j.coi.2012.12.006. Epub 2013 Jan 9. Curr Opin Immunol. 2013. PMID: 23313533 Free PMC article. Review.
References
- Medzhitov R. Recognition of microorganisms and activation of the immune response. Nature. 2007;449:819–826. - PubMed
- Pichlmair A, Reis e Sousa C. Innate recognition of viruses. Immunity. 2007;27:370–383. - PubMed
- Alexopoulou L, Holt AC, Medzhitov R, Flavell RA. Recognition of double-stranded RNA and activation of NF-[kappa]B by Toll-like receptor 3. Nature. 2001;413:732–738. - PubMed
- O’Neill LA, Bowie AG. The family of five: TIR-domain-containing adaptors in Toll-like receptor signalling. Nat Rev Immunol. 2007;7:353–364. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R37 AI067497/AI/NIAID NIH HHS/United States
- R01 AI067497/AI/NIAID NIH HHS/United States
- R01 AI083641/AI/NIAID NIH HHS/United States
- R56 AI067497/AI/NIAID NIH HHS/United States
- AI067497/AI/NIAID NIH HHS/United States
- ImNIH/Intramural NIH HHS/United States
- R01 HL093262/HL/NHLBI NIH HHS/United States
- R01 AI083713/AI/NIAID NIH HHS/United States
- AI083713/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous