A widespread chromosomal inversion polymorphism contributes to a major life-history transition, local adaptation, and reproductive isolation - PubMed (original) (raw)
A widespread chromosomal inversion polymorphism contributes to a major life-history transition, local adaptation, and reproductive isolation
David B Lowry et al. PLoS Biol. 2010.
Erratum in
- PLoS Biol. 2012 Jan;10(1):10.1371/annotation/caa1b7dd-9b6d-44db-b6ce-666954903625
Abstract
The role of chromosomal inversions in adaptation and speciation is controversial. Historically, inversions were thought to contribute to these processes either by directly causing hybrid sterility or by facilitating the maintenance of co-adapted gene complexes. Because inversions suppress recombination when heterozygous, a recently proposed local adaptation mechanism predicts that they will spread if they capture alleles at multiple loci involved in divergent adaptation to contrasting environments. Many empirical studies have found inversion polymorphisms linked to putatively adaptive phenotypes or distributed along environmental clines. However, direct involvement of an inversion in local adaptation and consequent ecological reproductive isolation has not to our knowledge been demonstrated in nature. In this study, we discovered that a chromosomal inversion polymorphism is geographically widespread, and we test the extent to which it contributes to adaptation and reproductive isolation under natural field conditions. Replicated crosses between the prezygotically reproductively isolated annual and perennial ecotypes of the yellow monkeyflower, Mimulus guttatus, revealed that alternative chromosomal inversion arrangements are associated with life-history divergence over thousands of kilometers across North America. The inversion polymorphism affected adaptive flowering time divergence and other morphological traits in all replicated crosses between four pairs of annual and perennial populations. To determine if the inversion contributes to adaptation and reproductive isolation in natural populations, we conducted a novel reciprocal transplant experiment involving outbred lines, where alternative arrangements of the inversion were reciprocally introgressed into the genetic backgrounds of each ecotype. Our results demonstrate for the first time in nature the contribution of an inversion to adaptation, an annual/perennial life-history shift, and multiple reproductive isolating barriers. These results are consistent with the local adaptation mechanism being responsible for the distribution of the two inversion arrangements across the geographic range of M. guttatus and that locally adaptive inversion effects contribute directly to reproductive isolation. Such a mechanism may be partially responsible for the observation that closely related species often differ by multiple chromosomal rearrangements.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
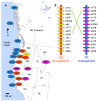
Figure 1. Geographic distribution of the chromosomal inversion.
(A) Map of western North America with the locations of populations of coastal perennials (blue), inland annuals (orange), and inland perennials (purple), as well as obligate self-fertilizing species M. nasutus (yellow). (B) Marker order of the AN and PE inversion arrangements along linkage group eight. Inland annuals and M. nasutus had the AN arrangement while coastal and inland perennials all had the PE arrangement.
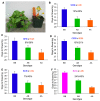
Figure 2. Replicated effect of the inversion locus.
(A) F2 progeny with parental ecotypic phenotypes, from a cross between the SWB (coastal perennial) and LMC (inland annual) populations. (B–E) Effect of the inversion on flowering time in four independently derived F2 mapping populations created through crosses between independent inland annual and coastal perennial populations. (F) Effects of the inversion on flowering time in cross between inland annual and inland perennial populations. The mean flowering times (±1 SE) of F2s that were homozygous for the AN arrangement (AA), heterozygous (AB), and homozygous for the PE arrangement (BB) at Micro6046 are indicated. The percentage of F2 variance/parental divergence explained by the inversion is presented above each bar graph. Note: _y_-axes do not originate at zero.
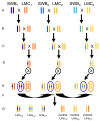
Figure 3. Breeding design for creation of backcross introgression lines.
Crossing design for production of backcross lines with the LMC (shades of yellow/orange) genetic background. Breeding of plants with SWB (shades of blue) genetic background not shown. Note that the size of introgressed region around the inversion should vary among lines due to different recombination locations during breeding. Different shades are used to indicate that original parental inbred lines have a unique genetic composition. (A) Three pairs of independently derived inbred LMC and SWB lines were crossed to create F1 progeny. (B) F1s backcrossed to parental inbred lines. (C) Marker-assisted selection used for four generations of backcrossing to move the inversion into alternate genetic backgrounds. (D) Heterozygous lines were self-fertilized. (E) Backcross lines that are homozygous with (blue oval) and without (orange oval) the introgressed inversion arrangements were selected for further breeding. (F) Intercrosses conducted among the three independent groups to create outbred backcross lines with and without the introgressed inverted region. (G) Backcrossed lines now ready to be planted into field reciprocal transplant experiment.
Figure 4. Effects of the inversion locus across field sites in different genetic backgrounds.
(A) Proportion of plants surviving to flower and (B) expected fitness produced per plants across field sites. Values plotted are maximum likelihood estimates ±1 SE calculated with ASTER. (C) Cumulative proportion of plants surviving to flower and (D) expected fitness per individual at the inland field site. Survival over time at the (E) coastal perennial and (F) inland annual field sites. Parental lines: yellow, inland annual parent; blue, coastal perennial parent. Backcross lines: orange, PE arrangement in annual genetic background; red, AN arrangement in annual genetic background; green, AN arrangement in perennial genetic background; pink, PE arrangement in perennial genetic background.
Similar articles
- Divergent population structure and climate associations of a chromosomal inversion polymorphism across the Mimulus guttatus species complex.
Oneal E, Lowry DB, Wright KM, Zhu Z, Willis JH. Oneal E, et al. Mol Ecol. 2014 Jun;23(11):2844-60. doi: 10.1111/mec.12778. Mol Ecol. 2014. PMID: 24796267 Free PMC article. - Dissecting the role of a large chromosomal inversion in life history divergence throughout the Mimulus guttatus species complex.
Coughlan JM, Willis JH. Coughlan JM, et al. Mol Ecol. 2019 Mar;28(6):1343-1357. doi: 10.1111/mec.14804. Epub 2018 Aug 11. Mol Ecol. 2019. PMID: 30028906 - Pooled ecotype sequencing reveals candidate genetic mechanisms for adaptive differentiation and reproductive isolation.
Gould BA, Chen Y, Lowry DB. Gould BA, et al. Mol Ecol. 2017 Jan;26(1):163-177. doi: 10.1111/mec.13881. Epub 2016 Nov 3. Mol Ecol. 2017. PMID: 27747958 - Eco-Evolutionary Genomics of Chromosomal Inversions.
Wellenreuther M, Bernatchez L. Wellenreuther M, et al. Trends Ecol Evol. 2018 Jun;33(6):427-440. doi: 10.1016/j.tree.2018.04.002. Epub 2018 May 3. Trends Ecol Evol. 2018. PMID: 29731154 Review. - Inversions and parallel evolution.
Westram AM, Faria R, Johannesson K, Butlin R, Barton N. Westram AM, et al. Philos Trans R Soc Lond B Biol Sci. 2022 Aug;377(1856):20210203. doi: 10.1098/rstb.2021.0203. Epub 2022 Jun 13. Philos Trans R Soc Lond B Biol Sci. 2022. PMID: 35694747 Free PMC article. Review.
Cited by
- Finding the Genomic Basis of Local Adaptation: Pitfalls, Practical Solutions, and Future Directions.
Hoban S, Kelley JL, Lotterhos KE, Antolin MF, Bradburd G, Lowry DB, Poss ML, Reed LK, Storfer A, Whitlock MC. Hoban S, et al. Am Nat. 2016 Oct;188(4):379-97. doi: 10.1086/688018. Epub 2016 Aug 15. Am Nat. 2016. PMID: 27622873 Free PMC article. Review. - The color genes of speciation in plants.
Ortiz-Barrientos D. Ortiz-Barrientos D. Genetics. 2013 May;194(1):39-42. doi: 10.1534/genetics.113.150466. Genetics. 2013. PMID: 23633142 Free PMC article. No abstract available. - Patterns of genetic variation across inversions: geographic variation in the In(2L)t inversion in populations of Drosophila melanogaster from eastern Australia.
Kennington WJ, Hoffmann AA. Kennington WJ, et al. BMC Evol Biol. 2013 May 20;13:100. doi: 10.1186/1471-2148-13-100. BMC Evol Biol. 2013. PMID: 23688159 Free PMC article. - Annelid Comparative Genomics and the Evolution of Massive Lineage-Specific Genome Rearrangement in Bilaterians.
Lewin TD, Liao IJ, Luo YJ. Lewin TD, et al. Mol Biol Evol. 2024 Sep 4;41(9):msae172. doi: 10.1093/molbev/msae172. Mol Biol Evol. 2024. PMID: 39141777 Free PMC article. - Five anthocyanin polymorphisms are associated with an R2R3-MYB cluster in Mimulus guttatus (Phrymaceae).
Lowry DB, Sheng CC, Lasky JR, Willis JH. Lowry DB, et al. Am J Bot. 2012 Jan;99(1):82-91. doi: 10.3732/ajb.1100285. Epub 2011 Dec 20. Am J Bot. 2012. PMID: 22186184 Free PMC article.
References
- Stebbins G. L. The inviability, weakness, and sterility of interspecific hybrids. Adv Genet. 1958;9:147–215. - PubMed
- Mayr E. Speciation and macroevolution. Evolution. 1982;36:1119–1132. - PubMed
- Clausen J. Stages in the evolution of plant species. Ithaca, NY: Cornell University Press; 1951.
- Dobzhansky T. Genetics of the evolutionary process. New York: Columbia University Press; 1970.
- White M. J. D. Modes of speciation. San Francisco: W.H. Freeman; 1978.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources